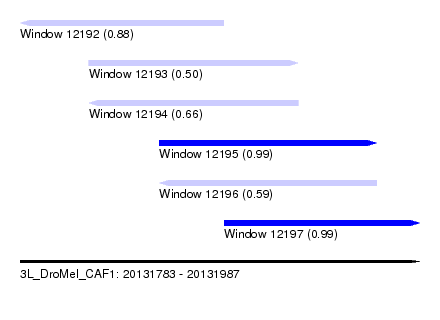
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,131,783 – 20,131,987 |
| Length | 204 |
| Max. P | 0.991647 |

| Location | 20,131,783 – 20,131,887 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.88 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20131783 104 - 23771897 AUUUGAAAAGCCAACAGCUGUGAGGCAAAAA-------AAAAACAAAUAUAUGCAUA----UACGUACAUAUAUAUUCAAGUGCAAAAAGCUGGCGUUGCUA-AUAGAAAUAAAUA (((((((..(((.((....))..))).....-------........(((((((.((.----...)).))))))).)))))))((((...(....).))))..-............. ( -16.40) >DroSec_CAF1 45961 111 - 1 AUUUGAAAAGCCAACAGCUGUGCGGCAAAAAAAAAAAAAAAAACAAAUAUAUGCAUA----UACGUACAUAUAUAUUCAAGUGCAAAAAGCUGGCGUUGCUA-AUAGAAAUAAAUA (((((((..(((.((....))..)))....................(((((((.((.----...)).))))))).)))))))((((...(....).))))..-............. ( -16.80) >DroSim_CAF1 46145 105 - 1 AUUUGAAAAGCCAACAGCUGUGCGGCAAAAAA------AAAAACAAAUAUAUGCAUA----UACGUACAUAUAUAUUCAAGUGCAAAAAGCUGGCGUUGCUA-AUAGAAAUAAAUA (((((((..(((.((....))..)))......------........(((((((.((.----...)).))))))).)))))))((((...(....).))))..-............. ( -16.80) >DroEre_CAF1 47985 112 - 1 AUUUGACAGGCUAACAGCUGUGCGGCAAAAAAA----AAAAAACAAAUAUAUGCAUAUCCAUACAUACAUAUAUAUUCAAGUGCAAAAAGCUGGCGUUGCUAGAUAGAAAUAAAUA .((((.(.(((...(((((.((((.........----......)..(((((((.((........)).)))))))........)))...))))).....))).).))))........ ( -16.36) >DroYak_CAF1 50368 102 - 1 AUUUGAAAGGCUAACAGCUGUGCGGCAAAAAAG----AAAAAACAAAUAUAUACAUA--------UAGAUAUAUAUUCAAGUGCAA-AAGCUGGCGUUGCUAAAUAGAAAUAAA-A ........(((...(((((.....(((......----........((((((((....--------....))))))))....)))..-.))))).....))).............-. ( -14.07) >consensus AUUUGAAAAGCCAACAGCUGUGCGGCAAAAAA_____AAAAAACAAAUAUAUGCAUA____UACGUACAUAUAUAUUCAAGUGCAAAAAGCUGGCGUUGCUA_AUAGAAAUAAAUA (((((((..(((.((....))..)))....................(((((((.((........)).))))))).)))))))((((...(....).))))................ (-14.22 = -14.46 + 0.24)

| Location | 20,131,818 – 20,131,925 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -18.38 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20131818 107 + 23771897 UUGAAUAUAUAUGUACGUA----UAUGCAUAUAUUUGUUUUU-------UUUUUGCCUCACAGCUGUUGGCUUUUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAA ......(((((((((....----..)))))))))...((((.-------((((((.....((((..((((....))))..((((((.....))).))))))).....)))))).)))) ( -22.10) >DroSec_CAF1 45996 114 + 1 UUGAAUAUAUAUGUACGUA----UAUGCAUAUAUUUGUUUUUUUUUUUUUUUUUGCCGCACAGCUGUUGGCUUUUCAAAUGACGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAA .......((((((((....----..))))))))(((((((((......((....((.((.((.(((..(((.((......)).)))...))).)))).)).)).....))))))))). ( -23.40) >DroSim_CAF1 46180 108 + 1 UUGAAUAUAUAUGUACGUA----UAUGCAUAUAUUUGUUUUU------UUUUUUGCCGCACAGCUGUUGGCUUUUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAA .......((((((((....----..))))))))(((((((((------...((.((.((.((.(((..(((.((......)).)))...))).)))).)).)).....))))))))). ( -23.90) >DroEre_CAF1 48021 114 + 1 UUGAAUAUAUAUGUAUGUAUGGAUAUGCAUAUAUUUGUUUUUU----UUUUUUUGCCGCACAGCUGUUAGCCUGUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAA .......(((((((((((....)))))))))))(((((((((.----....((.((.((.((.(((...(((........)))......))).)))).)).)).....))))))))). ( -28.90) >DroYak_CAF1 50402 105 + 1 UUGAAUAUAUAUCUA--------UAUGUAUAUAUUUGUUUUUU----CUUUUUUGCCGCACAGCUGUUAGCCUUUCAAAUGGCGC-UUUCAGGUGGCCGCUGAUCCACGAAAAUAAAA ....((((((((...--------.)))))))).(((((((((.----....((.((.((.((.(((..(((((.......)).))-)..))).)))).)).)).....))))))))). ( -23.90) >consensus UUGAAUAUAUAUGUACGUA____UAUGCAUAUAUUUGUUUUUU_____UUUUUUGCCGCACAGCUGUUGGCUUUUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAA .......((((((((..........))))))))(((((((((.......((...((.((.((.(((..(((((.......)).)))...))).)))).)).)).....))))))))). (-18.38 = -18.62 + 0.24)

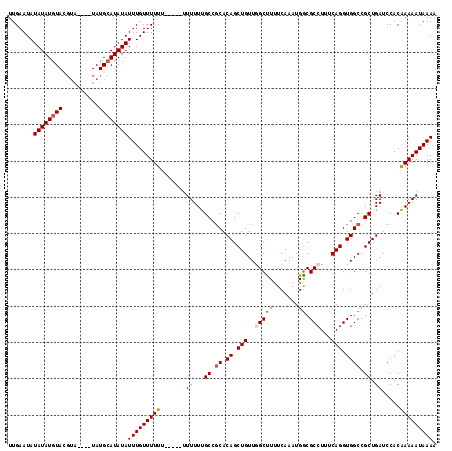
| Location | 20,131,818 – 20,131,925 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -22.11 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20131818 107 - 23771897 UUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGAAAAGCCAACAGCUGUGAGGCAAAAA-------AAAAACAAAUAUAUGCAUA----UACGUACAUAUAUAUUCAA ((((.(((((((....(((((((..((.....)).)))..(((......)))..))))....)))))))-------.))))...(((((((.((.----...)).)))))))...... ( -20.50) >DroSec_CAF1 45996 114 - 1 UUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGUCAUUUGAAAAGCCAACAGCUGUGCGGCAAAAAAAAAAAAAAAAACAAAUAUAUGCAUA----UACGUACAUAUAUAUUCAA ((((.(((((((.(..((((((...)).....(((.((....))...)))....))))..).))))))).))))..........(((((((.((.----...)).)))))))...... ( -23.10) >DroSim_CAF1 46180 108 - 1 UUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGAAAAGCCAACAGCUGUGCGGCAAAAAA------AAAAACAAAUAUAUGCAUA----UACGUACAUAUAUAUUCAA ((((.(((((((.(..(((((((..((.....)).)))..(((......)))..))))..).))))))).------))))....(((((((.((.----...)).)))))))...... ( -23.20) >DroEre_CAF1 48021 114 - 1 UUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGACAGGCUAACAGCUGUGCGGCAAAAAAA----AAAAAACAAAUAUAUGCAUAUCCAUACAUACAUAUAUAUUCAA ((((.(((((((.(..((((.(((........)))(((........))).....))))..).))))))).)----)))......(((((((.((........)).)))))))...... ( -26.10) >DroYak_CAF1 50402 105 - 1 UUUUAUUUUCGUGGAUCAGCGGCCACCUGAAA-GCGCCAUUUGAAAGGCUAACAGCUGUGCGGCAAAAAAG----AAAAAACAAAUAUAUACAUA--------UAGAUAUAUAUUCAA .....(((((........((.((((.(((...-..(((........)))...))).)).)).))......)----))))....((((((((....--------....))))))))... ( -17.64) >consensus UUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGAAAAGCCAACAGCUGUGCGGCAAAAAA_____AAAAAACAAAUAUAUGCAUA____UACGUACAUAUAUAUUCAA .....(((((((.(..(((((((..((.....)).)))..(((......)))..))))..).)))))))...............(((((((.((........)).)))))))...... (-16.90 = -17.38 + 0.48)



| Location | 20,131,854 – 20,131,965 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -30.42 |
| Energy contribution | -31.46 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20131854 111 + 23771897 UU-------UUUUUGCCUCACAGCUGUUGGCUUUUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAAACAACA ..-------.(((.(((...((((..((((....))))..((((((.....))).)))))))...............(((((((((.........)))))))))..))).)))..... ( -33.20) >DroSec_CAF1 46032 118 + 1 UUUUUUUUUUUUUUGCCGCACAGCUGUUGGCUUUUCAAAUGACGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAACAAACA ..............((((..((((....(((...((....))((((.....)))))))))))...............(((((((((.........))))))))).))))......... ( -33.90) >DroSim_CAF1 46216 112 + 1 UU------UUUUUUGCCGCACAGCUGUUGGCUUUUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAACAAACA ..------......((......))((((((((..(((..(((((((.....))).)))).)))..............(((((((((.........)))))))))..)))))))).... ( -35.70) >DroEre_CAF1 48061 114 + 1 UUU----UUUUUUUGCCGCACAGCUGUUAGCCUGUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCGAAAAACA ...----((((((.((((..((((.....(((........)))(((........))).))))...............(((((((((.........))))))))).)))).)))))).. ( -39.60) >DroYak_CAF1 50434 113 + 1 UUU----CUUUUUUGCCGCACAGCUGUUAGCCUUUCAAAUGGCGC-UUUCAGGUGGCCGCUGAUCCACGAAAAUAAAAGACACGCGGAAAAGAGACGCGUGUCUCCUGUCAAAAAACA ...----.((((((((.((.((.(((..(((((.......)).))-)..))).)))).))............(((..(((((((((.........)))))))))..))).)))))).. ( -31.90) >consensus UUU_____UUUUUUGCCGCACAGCUGUUGGCUUUUCAAAUGGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAAAAAACA ........(((((.((((..((((.....(((........)))(((........))).))))...............(((((((((.........))))))))).)))).)))))... (-30.42 = -31.46 + 1.04)


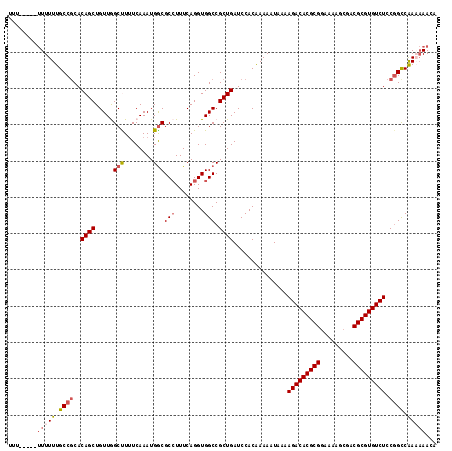
| Location | 20,131,854 – 20,131,965 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20131854 111 - 23771897 UGUUGUUUGGCCGGAGACACGCGUCGCUUUUCCGCGUGUCUUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGAAAAGCCAACAGCUGUGAGGCAAAAA-------AA .((((((((((((((((((((((.........))))))))))......(((.....))))))))).......(((............))))))))).............-------.. ( -34.40) >DroSec_CAF1 46032 118 - 1 UGUUUGUUGGCCGGAGACACGCGUCGCUUUUCCGCGUGUCUUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGUCAUUUGAAAAGCCAACAGCUGUGCGGCAAAAAAAAAAAAAA ....(((((((((((((((((((.........))))))))))......(((.....))))))))........(((.((....))...)))))))(((....))).............. ( -35.50) >DroSim_CAF1 46216 112 - 1 UGUUUGUUGGCCGGAGACACGCGUCGCUUUUCCGCGUGUCUUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGAAAAGCCAACAGCUGUGCGGCAAAAAA------AA ....(((((((.(((((((((((.........)))))))))))..(((..((((.((((.(....)))))......))))..)))..)))))))(((....)))......------.. ( -36.50) >DroEre_CAF1 48061 114 - 1 UGUUUUUCGGCCGGAGACACGCGUCGCUUUUCCGCGUGUCUUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGACAGGCUAACAGCUGUGCGGCAAAAAAA----AAA ..(((((..((((((((((((((.........))))))))))..............((((.(((........)))(((........))).....))))..))))..)))))----... ( -38.60) >DroYak_CAF1 50434 113 - 1 UGUUUUUUGACAGGAGACACGCGUCUCUUUUCCGCGUGUCUUUUAUUUUCGUGGAUCAGCGGCCACCUGAAA-GCGCCAUUUGAAAGGCUAACAGCUGUGCGGCAAAAAAG----AAA (.(((((((..((((((((((((.........))))))))))))....((((....((((.((.........-))(((........))).....)))).))))))))))).----).. ( -32.70) >consensus UGUUUUUUGGCCGGAGACACGCGUCGCUUUUCCGCGUGUCUUUUAUUUUUGUGGAUCAGCGGCCACCUGAAAGGCGCCAUUUGAAAAGCCAACAGCUGUGCGGCAAAAAA_____AAA ............(((((((((((.........)))))))))))..(((((((.(..(((((((..((.....)).)))..(((......)))..))))..).)))))))......... (-30.12 = -30.32 + 0.20)


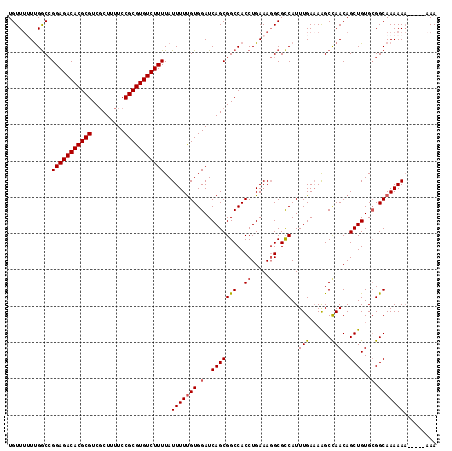
| Location | 20,131,887 – 20,131,987 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -22.84 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20131887 100 + 23771897 GGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAAACAACAAUAACAACAACCCCUAUUUCUU ((.(((.....)))(((((.((.....))........(((((((((.........))))))))).)))))....................))........ ( -26.30) >DroSec_CAF1 46072 100 + 1 GACGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAACAAACAAUAACAACAACGACUAUGUCCU ((((((.....)))(((((.((.....))........(((((((((.........))))))))).))))).........................))).. ( -28.20) >DroSim_CAF1 46250 100 + 1 GGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAACAAACAAUAACAACAACGACUAUUUCCU ((.(((.....)))(((((.((.....))........(((((((((.........))))))))).)))))...........................)). ( -26.10) >DroEre_CAF1 48097 100 + 1 GGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCGAAAAACAAUAACAACAAUGGCUAUUUCCU ((((((.....)))(((((.((.....))........(((((((((.........))))))))).)))))....................)))....... ( -28.50) >DroYak_CAF1 50470 99 + 1 GGCGC-UUUCAGGUGGCCGCUGAUCCACGAAAAUAAAAGACACGCGGAAAAGAGACGCGUGUCUCCUGUCAAAAAACAAUAACCACAACCGCUAUUUCCU ((((.-((((..((((........))))))))(((..(((((((((.........)))))))))..)))....................))))....... ( -23.80) >consensus GGCGCCUUUCAGGUGGCCGCUGAUCCACAAAAAUAAAAGACACGCGGAAAAGCGACGCGUGUCUCCGGCCAAAAAACAAUAACAACAACGACUAUUUCCU (((.........((((........)))).........(((((((((.........)))))))))...))).............................. (-22.84 = -22.92 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:49 2006