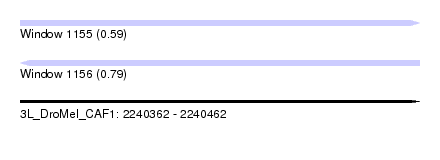
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,240,362 – 2,240,462 |
| Length | 100 |
| Max. P | 0.792017 |

| Location | 2,240,362 – 2,240,462 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -18.27 |
| Energy contribution | -20.60 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
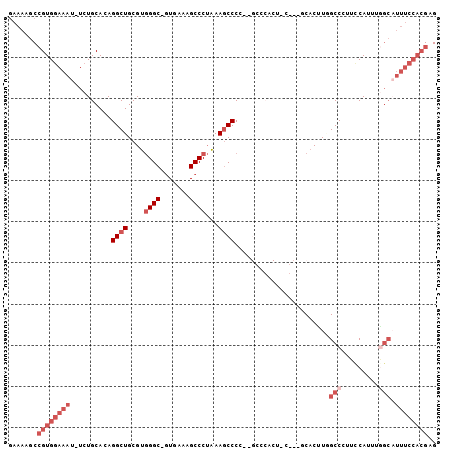
>3L_DroMel_CAF1 2240362 100 + 23771897 GAAAAGCCGUGGAAAU-UCUGCACAGGCUGCGUGGGC-GUGAAAGCCCUAAAGCCCC--GCCCACUCUGCCGCACUUGGCCAUUUCAUUUGGCAUUUCCACGAG .......(((((((((-..(((...(((.(.((((((-(.(...((......)).))--)))))).).))))))....((((.......))))))))))))).. ( -38.90) >DroSec_CAF1 1265 88 + 1 CAAAAGCCGUGGAAAU-UCUGCACAGGCUGCGUGGGC-GUGAAAGCCCUAAAGCCCC--------------CCACCUUGCCCUUCCAUUGGGCAUUUCCACGAG .......((((((((.-........((((....((((-......))))...))))..--------------......(((((.......))))))))))))).. ( -34.90) >DroSim_CAF1 3510 88 + 1 GAAAAGCCGUGGAAAU-UCUGCACAGGCUGCGUGGGC-GUGAAAGCCCUAAAGCCCC--------------CCACUUUGCCCUUCCAUUGAGCAUUUCCACGAG .......((((((((.-..(((...((((....((((-......))))...))))..--------------...(..((......))..).))))))))))).. ( -28.10) >DroEre_CAF1 3987 100 + 1 GAAAAGCCGUGGAAAU-UCUGCACAGGCUGCGUGGGC-UUGAAAGCCCUAAAGCCCC--GCCCACUCCGCCGCACUUGGCCCUUCCAUUUGGCAUUUCCACGAG .......(((((((((-...((...((((....((((-(....)))))...))))..--)).......((((....(((.....)))..))))))))))))).. ( -34.60) >DroYak_CAF1 4161 100 + 1 GAAAAGCCGUGGAAAU-UCUGCACAGGCUGCGUGGGC-GUGAAAGCCCUAAAGCCCC--GCCCACUUCGCCGCACUUGGCCCUUCCAUUUGGCUUUUCCACGAG ((((((((((((((..-...((...((((....((((-......))))...))))..--)).......((((....))))..))))))..))))))))...... ( -37.00) >DroAna_CAF1 6812 86 + 1 GAAAAGCGGUGAAAAUGUCUGCAAAGGCUGCGUAGGCGGUGAAAGCCCAGAAUCCCUCCGACCACCAC----CCCUUAGCCCGCACA--------------GAG .....((((.......((((....)))).((..(((.((((........................)))----))))..))))))...--------------... ( -19.16) >consensus GAAAAGCCGUGGAAAU_UCUGCACAGGCUGCGUGGGC_GUGAAAGCCCUAAAGCCCC__GCCCACU_C___GCACUUGGCCCUUCCAUUUGGCAUUUCCACGAG .......((((((((..........((((....((((.......))))...)))).......................(((.........))).)))))))).. (-18.27 = -20.60 + 2.33)

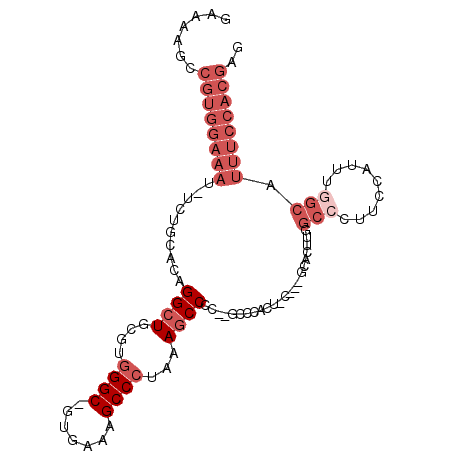
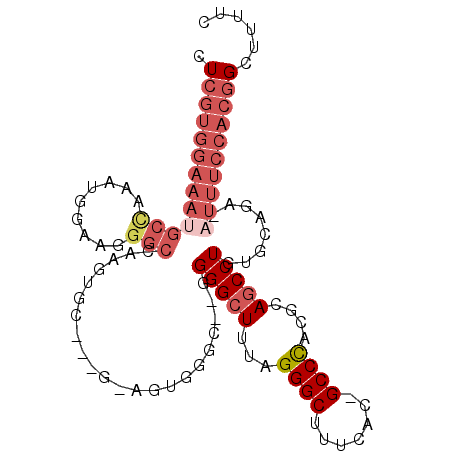
| Location | 2,240,362 – 2,240,462 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -20.47 |
| Energy contribution | -22.55 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2240362 100 - 23771897 CUCGUGGAAAUGCCAAAUGAAAUGGCCAAGUGCGGCAGAGUGGGC--GGGGCUUUAGGGCUUUCAC-GCCCACGCAGCCUGUGCAGA-AUUUCCACGGCUUUUC .((((((((((((((.......))))...(..((((...((((((--(.((((....))))....)-))))))...))).)..)...-))))))))))...... ( -44.40) >DroSec_CAF1 1265 88 - 1 CUCGUGGAAAUGCCCAAUGGAAGGGCAAGGUGG--------------GGGGCUUUAGGGCUUUCAC-GCCCACGCAGCCUGUGCAGA-AUUUCCACGGCUUUUG .((((((((((((((.......))))..(((((--------------..((((....))))..).)-)))...(((.....)))...-))))))))))...... ( -35.60) >DroSim_CAF1 3510 88 - 1 CUCGUGGAAAUGCUCAAUGGAAGGGCAAAGUGG--------------GGGGCUUUAGGGCUUUCAC-GCCCACGCAGCCUGUGCAGA-AUUUCCACGGCUUUUC .((((((((((.((........((((...(((.--------------.((.(.....).))..)))-))))..(((.....))))).-))))))))))...... ( -32.20) >DroEre_CAF1 3987 100 - 1 CUCGUGGAAAUGCCAAAUGGAAGGGCCAAGUGCGGCGGAGUGGGC--GGGGCUUUAGGGCUUUCAA-GCCCACGCAGCCUGUGCAGA-AUUUCCACGGCUUUUC .(((((((((((((.........)))...(..((((...((((((--.(((((....)))))....-))))))...))).)..)...-))))))))))...... ( -42.10) >DroYak_CAF1 4161 100 - 1 CUCGUGGAAAAGCCAAAUGGAAGGGCCAAGUGCGGCGAAGUGGGC--GGGGCUUUAGGGCUUUCAC-GCCCACGCAGCCUGUGCAGA-AUUUCCACGGCUUUUC ......((((((((...((((((..(...(..((((...((((((--(.((((....))))....)-))))))...))).)..).).-.)))))).)))))))) ( -44.40) >DroAna_CAF1 6812 86 - 1 CUC--------------UGUGCGGGCUAAGGG----GUGGUGGUCGGAGGGAUUCUGGGCUUUCACCGCCUACGCAGCCUUUGCAGACAUUUUCACCGCUUUUC .((--------------((((.(((((...((----(((((((((((((...))))))....)))))))))....))))).))))))................. ( -30.70) >consensus CUCGUGGAAAUGCCAAAUGGAAGGGCCAAGUGC___G_AGUGGGC__GGGGCUUUAGGGCUUUCAC_GCCCACGCAGCCUGUGCAGA_AUUUCCACGGCUUUUC .(((((((((((((.........)))......................(((((...((((.......))))....)))))........))))))))))...... (-20.47 = -22.55 + 2.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:15 2006