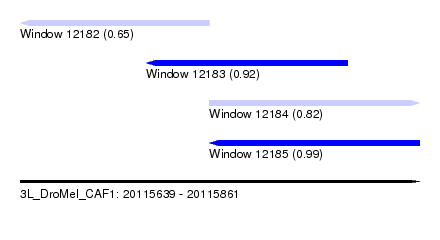
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,115,639 – 20,115,861 |
| Length | 222 |
| Max. P | 0.994701 |

| Location | 20,115,639 – 20,115,744 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.73 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -5.34 |
| Energy contribution | -5.06 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20115639 105 - 23771897 AAAAACGA-----AAAAAACUUGCUUGGACAUAUUGCUGCU------GUGCAUUCU----CUUCAACCAAUUGGAGAAGAAUGAAAUCGAGGGGAAAACUAAGACAAGGCAACGCGUUAU ...((((.-----......(((((((((......(((....------..)))((((----((((..((....))....((......))))))))))..))))).))))......)))).. ( -25.42) >DroSec_CAF1 29446 99 - 1 AGGAACAAAAAAAAAAAAACAUGCUU------GUUGCUGUAGCGGAAAUGCAUUUU----CUUCAACCAUUCGAAGAAGAAUGAAAUCGAGGGAAAAACUAAAACUAAA----------- .....................(((..------.((((....))))....)))((((----((((...(((((......))))).....)))))))).............----------- ( -16.00) >DroSim_CAF1 29918 115 - 1 AGGAACGAAAA---AAAAACAUGCUUGGACAUGUUGCUGUAGCGGAAAUGCAUUUUCUUUCUUCAAGCAAUUGAAGAAGAAUGAAAUCGAGGGGAAAACUAAGACAAAGCUU--CGUUAU ...((((((..---...((((((......))))))(((...(((....))).((..((((((((((....))))))).((......)))))..))............)))))--)))).. ( -26.00) >DroEre_CAF1 31982 97 - 1 AAGAACGAAA------GAACAUACUUGGACAUAUUGCUAUGGGGGAAAUGUAUUUU----CUUAAACCAAUUGAAGAAGAGUGAAAUCGAUUGGAAAACUAACCCAA------------- .....(....------)......................(((((((((.....)))----))....((((((((............))))))))........)))).------------- ( -17.20) >DroYak_CAF1 33362 105 - 1 AAGAACGAAA------AAACAUAUUUCGACAUAUUGCUGUAUUGU--AUGUAU-UU----CUUCAACCAAUUGAAGAAGAAUAAAAUCGAAUAGAAAACUAAGACAAAGUUA--UGUUUU .........(------(((((((.((((((((((.........))--)))).(-((----((((((....)))))))))........))))(((....))).........))--)))))) ( -17.50) >consensus AAGAACGAAA____AAAAACAUGCUUGGACAUAUUGCUGUAGCGGAAAUGCAUUUU____CUUCAACCAAUUGAAGAAGAAUGAAAUCGAGGGGAAAACUAAGACAAAG______GUU_U ..................................................((((((....((((((....)))))).))))))..................................... ( -5.34 = -5.06 + -0.28)


| Location | 20,115,709 – 20,115,821 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20115709 112 - 23771897 AAAUGCGGCAAACAUGAAAUG--CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUUGAGCUUUAAAAAACGA-----AAAAAACUUGCUUGGACAUAUUGCUGC ....(((((((..(((..(((--((((.........)))))))..........((((........))))(..(((............-----.........)))..).))).))))))) ( -25.10) >DroSec_CAF1 29511 105 - 1 AAAUGCGGCAAA-----AAUG--CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAGCUU-AAGGAACAAAAAAAAAAAAACAUGCUU------GUUGCUGU ....(((((((.-----.(((--((((.........)))))))..........((((........)))).(((((..-.......................))))------)))))))) ( -23.01) >DroSim_CAF1 29996 108 - 1 AAAUGCGGCAAA-----AAUG--CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAACUU-AAGGAACGAAAA---AAAAACAUGCUUGGACAUGUUGCUGU ....(((((...-----.(((--((((.........)))))))..(((((.((..((((..........(((..(..-..)...)))...---.......))))..)).)))))))))) ( -22.75) >DroEre_CAF1 32045 106 - 1 AAAUGCG-CAAA-----CAUGAAAUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAACCU-AAAGAACGAAA------GAACAUACUUGGACAUAUUGCUAU .......-....-----......((((((((....))))))))...........((((((((...((((((......-...)).(....------).......))))...)))))))). ( -22.10) >DroYak_CAF1 33433 107 - 1 AAAUGCGGCAAA-----CAUGAAAUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAUUCGAGCUU-AAAGAACGAAA------AAACAUAUUUCGACAUAUUGCUGU ....(((((((.-----.(((..((((((((....)))))))).((((((.((((((........)).((((..(..-...)..)))))------))).))))))...))).))))))) ( -23.80) >consensus AAAUGCGGCAAA_____AAUG__CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAGCUU_AAAGAACGAAA____AAAAACAUGCUUGGACAUAUUGCUGU ....(((((((.............(((((((....)))))))...........((((........))))...........................................))))))) (-16.72 = -17.16 + 0.44)


| Location | 20,115,744 – 20,115,861 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -11.49 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20115744 117 + 23771897 UAAAGCUCAAUUGGGUUUAUUGCCAAAAUAUAUUUUAUGCUGCAGUGGCGCAGCAG--CAUUUCAUGUUUGCCG-CAUUUUGGCCAAUUUAUAGCAAUUGAGUUUUCUUUAUCGAGUUGU .((((((((((((.((..(((((((((((........((((((......))))))(--((.........)))..-.))))))).))))..))..)))))))))))).............. ( -34.80) >DroSec_CAF1 29545 110 + 1 U-AAGCUCGAUUGGGUUUAUUGCCAAAAUAUAUUUUAUGCUGCAGUGGCGCAGCAG--CAUU-----UUUGCCG-CAUUUUGGCCAAUUUAUUGCAAUUGAGUU-UCUUUAUCGAGUUGU .-((((((((((((((..(((((((((((........((((((......))))))(--((..-----..)))..-.))))))).))))..))).))))))))))-).............. ( -34.20) >DroSim_CAF1 30033 110 + 1 U-AAGUUCGAUUGGGUUUAUUGCCAAAAUAUAUUUUAUGCUGCAGUGGCGCAGCAG--CAUU-----UUUGCCG-CAUUUUGGCUAAUUUAUUGCAAUUGAGUU-UCUUUAUCGAGUUGU .-...(((((((((((..(((((((((((........((((((......))))))(--((..-----..)))..-.)))))))).)))..))).))))))))..-............... ( -29.20) >DroEre_CAF1 32079 112 + 1 U-AGGUUCGAUUGGGUUUAUUGCCAAAAUAUAUUUUAUGCUGCAGUGGCGCAGCAUUUCAUG-----UUUG-CG-CAUUUUAGUCAAUUUAUUGCUAUUGAGUUUUCUUUAUCGAGUUUU .-(..((((((..((.....(((((((.........(((((((......)))))))......-----))))-.)-))......(((((........))))).....))..))))))..). ( -24.76) >DroYak_CAF1 33467 113 + 1 U-AAGCUCGAAUGGGUUUAUUGCCAAAAUAUAUUUUAUGCUGCAGUGGCGCAGCAUUUCAUG-----UUUGCCG-CAUUUUAGUCAAUUUAUUGCCAUUGAGUUUUCUUUAUCGAGUUUU .-((((((((...(((.....)))..............(((.(((((((((.(((.......-----..))).)-)....(((.....)))..))))))))))........)))))))). ( -27.70) >DroAna_CAF1 26632 96 + 1 U-AAGCCAGC--UAGCCCAUUUCCUGGCCACAUUAUAUGCUCCACUGGA--AGC-----------------CCGAAAACUUAACCCAUUUCAUGCCACCCAGUUCUGGUUAUAAA--UUG .-.((((((.--..(((........)))...............(((((.--.((-----------------..((((..........))))..))...))))).)))))).....--... ( -16.90) >consensus U_AAGCUCGAUUGGGUUUAUUGCCAAAAUAUAUUUUAUGCUGCAGUGGCGCAGCAG__CAUU_____UUUGCCG_CAUUUUAGCCAAUUUAUUGCAAUUGAGUUUUCUUUAUCGAGUUGU ...(((((((...(((.....))).............((((((......))))))..........................................)))))))................ (-11.49 = -12.18 + 0.70)

| Location | 20,115,744 – 20,115,861 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -11.14 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20115744 117 - 23771897 ACAACUCGAUAAAGAAAACUCAAUUGCUAUAAAUUGGCCAAAAUG-CGGCAAACAUGAAAUG--CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUUGAGCUUUA .........(((((....((((((((..........(((((((((-.((((.........))--))((((((....))))))........)))))))))........))))))))))))) ( -29.97) >DroSec_CAF1 29545 110 - 1 ACAACUCGAUAAAGA-AACUCAAUUGCAAUAAAUUGGCCAAAAUG-CGGCAAA-----AAUG--CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAGCUU-A ....((((((.....-....(((((......)))))(((((((((-.((((..-----..))--))((((((....))))))........)))))))))..........))))))...-. ( -32.10) >DroSim_CAF1 30033 110 - 1 ACAACUCGAUAAAGA-AACUCAAUUGCAAUAAAUUAGCCAAAAUG-CGGCAAA-----AAUG--CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAACUU-A .....(((((.....-.....((((......)))).(((((((((-.((((..-----..))--))((((((....))))))........)))))))))..........)))))....-. ( -26.70) >DroEre_CAF1 32079 112 - 1 AAAACUCGAUAAAGAAAACUCAAUAGCAAUAAAUUGACUAAAAUG-CG-CAAA-----CAUGAAAUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAACCU-A .....(((((...((....))...........((((.((((((((-..-((..-----..))..((((((((....))))))))......)))))))))))).......)))))....-. ( -20.40) >DroYak_CAF1 33467 113 - 1 AAAACUCGAUAAAGAAAACUCAAUGGCAAUAAAUUGACUAAAAUG-CGGCAAA-----CAUGAAAUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAUUCGAGCUU-A ....(((((....((....)).((((......((((.((((((((-.(.....-----).....((((((((....))))))))......))))))))))))....)))))))))...-. ( -24.10) >DroAna_CAF1 26632 96 - 1 CAA--UUUAUAACCAGAACUGGGUGGCAUGAAAUGGGUUAAGUUUUCGG-----------------GCU--UCCAGUGGAGCAUAUAAUGUGGCCAGGAAAUGGGCUA--GCUGGCUU-A ...--.......((((.((((((.(((.(((((..........))))).-----------------)))--)))))).............(((((........)))))--.))))...-. ( -24.20) >consensus AAAACUCGAUAAAGAAAACUCAAUUGCAAUAAAUUGGCCAAAAUG_CGGCAAA_____AAUG__CUGCUGCGCCACUGCAGCAUAAAAUAUAUUUUGGCAAUAAACCCAAUCGAGCUU_A .....(((((...((....))...........((((.((((((((....................(((((((....))))))).......)))))))))))).......)))))...... (-11.14 = -12.03 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:38 2006