| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,102,715 – 20,102,898 |
| Length | 183 |
| Max. P | 0.881698 |

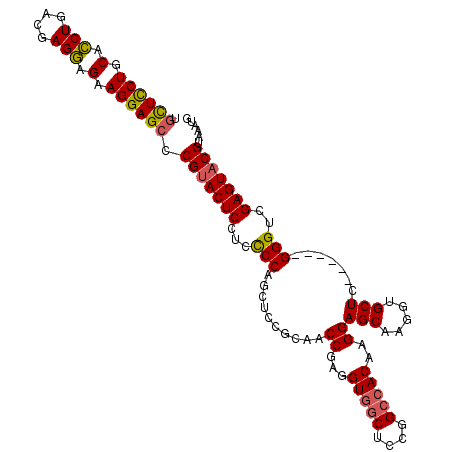
| Location | 20,102,715 – 20,102,821 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.04 |
| Mean single sequence MFE | -44.50 |
| Consensus MFE | -34.74 |
| Energy contribution | -34.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
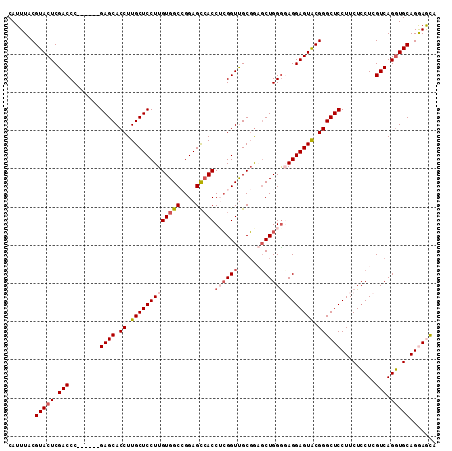
>3L_DroMel_CAF1 20102715 106 + 23771897 UACUCCUGCAACUGACGAGUAGAAGGAGCCCGUACUCCUCCCCAGCUUCGCAACCGAGGUGGCUCCGGCCACAAGGAGCAAGGUGCUC------GGGUCGAGUACGUAAAUG ..(((((.(.(((....))).).)))))..(((((((..(((.(((.((....((...(((((....)))))..)).....)).))).------)))..)))))))...... ( -41.60) >DroSec_CAF1 17067 106 + 1 UGCUCCUGCACCUGACGAGGAGAAGGAGCCCGUACUCCUGCCCAGCUCCGCAACCGUGGUUGCUCCGGCCACAAGGAGCAAGGUGCUC------GGGUCGAGUACGUAAAUG .((((((.(.(((....))).).)))))).(((((((..(((((((.((((....))))(((((((........)))))))...))).------)))).)))))))...... ( -48.20) >DroSim_CAF1 17465 106 + 1 UGCUCCUGCACCUGACGAGGAGAAGGAGCCCGUACUCCUGCCCAGCUCCGCAACCGUGGUGGCUCCGGCCACAAGGAGCAAGGUGCUC------GGGUCGAGUACGUAAAUG .((((((.(.(((....))).).)))))).(((((((..(((((((.((....((...(((((....)))))..)).....)).))).------)))).)))))))...... ( -48.70) >DroEre_CAF1 16779 106 + 1 UGUUUCUGCACCUGACGAGGAGAAGGAGCCCGCACUCCUCUCCAGCACGGCAACCCAGGUGGCUCCUGCCACAAGGAGCAAGGUGCUC------GGGUCGAGUACGUAAAUG ........((((((....(((((.((((......))))))))).....(....).))))))((((((......))))))...((((((------(...)))))))....... ( -44.00) >DroYak_CAF1 17273 112 + 1 UGUUUCUGCACCUGACGAGGAGAAGGAGCCCGUACUCCUCCCCAGGACCGCGACCGAGGUGACUCCUGCCACAAGGAGCAAGGUGCUCGAGUACGGGUCGAGUACGUAAAUG (((((((...((((..((((((..((...))...))))))..))))...(((...(((....))).)))....)))))))..((((((((.......))))))))....... ( -40.00) >consensus UGCUCCUGCACCUGACGAGGAGAAGGAGCCCGUACUCCUCCCCAGCUCCGCAACCGAGGUGGCUCCGGCCACAAGGAGCAAGGUGCUC______GGGUCGAGUACGUAAAUG .((((((.(.(((....))).).)))))).(((((((...(((..........((...(((((....)))))..))(((.....))).......)))..)))))))...... (-34.74 = -34.58 + -0.16)

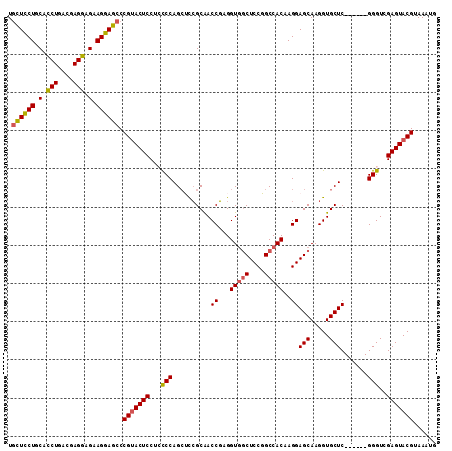
| Location | 20,102,715 – 20,102,821 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.04 |
| Mean single sequence MFE | -45.24 |
| Consensus MFE | -34.83 |
| Energy contribution | -36.31 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
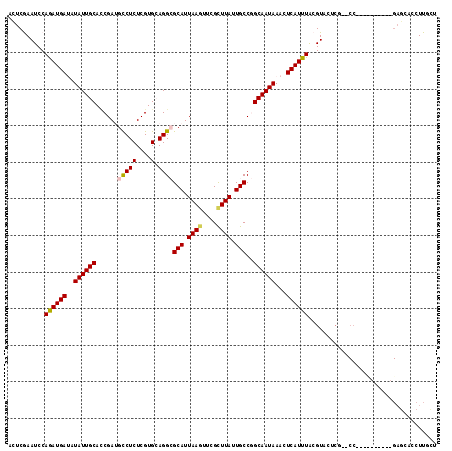
>3L_DroMel_CAF1 20102715 106 - 23771897 CAUUUACGUACUCGACCC------GAGCACCUUGCUCCUUGUGGCCGGAGCCACCUCGGUUGCGAAGCUGGGGAGGAGUACGGGCUCCUUCUACUCGUCAGUUGCAGGAGUA ......(((((((..(((------.(((...((((.((..(((((....)))))...))..)))).))).)))..))))))).((((((.(.(((....))).).)))))). ( -44.90) >DroSec_CAF1 17067 106 - 1 CAUUUACGUACUCGACCC------GAGCACCUUGCUCCUUGUGGCCGGAGCAACCACGGUUGCGGAGCUGGGCAGGAGUACGGGCUCCUUCUCCUCGUCAGGUGCAGGAGCA ......(((((((.....------((((.....))))....((.((((.(((((....)))))....)))).)).))))))).((((((.(.(((....))).).)))))). ( -47.10) >DroSim_CAF1 17465 106 - 1 CAUUUACGUACUCGACCC------GAGCACCUUGCUCCUUGUGGCCGGAGCCACCACGGUUGCGGAGCUGGGCAGGAGUACGGGCUCCUUCUCCUCGUCAGGUGCAGGAGCA ......(((((((..(((------((((.....))))...((..(((.((((.....)))).))).)).)))...))))))).((((((.(.(((....))).).)))))). ( -46.60) >DroEre_CAF1 16779 106 - 1 CAUUUACGUACUCGACCC------GAGCACCUUGCUCCUUGUGGCAGGAGCCACCUGGGUUGCCGUGCUGGAGAGGAGUGCGGGCUCCUUCUCCUCGUCAGGUGCAGAAACA .......((((..(((..------.(((((...((.(((.(((((....)))))..)))..)).)))))((((((((((....))))).)))))..)))..))))....... ( -44.70) >DroYak_CAF1 17273 112 - 1 CAUUUACGUACUCGACCCGUACUCGAGCACCUUGCUCCUUGUGGCAGGAGUCACCUCGGUCGCGGUCCUGGGGAGGAGUACGGGCUCCUUCUCCUCGUCAGGUGCAGAAACA .......((((..((((((.((.((((.((((((((......)))))).))...)))))))).))))((((.((((((...((....)).)))))).))))))))....... ( -42.90) >consensus CAUUUACGUACUCGACCC______GAGCACCUUGCUCCUUGUGGCCGGAGCCACCUCGGUUGCGGAGCUGGGGAGGAGUACGGGCUCCUUCUCCUCGUCAGGUGCAGGAGCA .......(((((.(((........((((.((.(((((((((((((....)))))((((((......)))))))))))))).)))))).........))).)))))....... (-34.83 = -36.31 + 1.48)

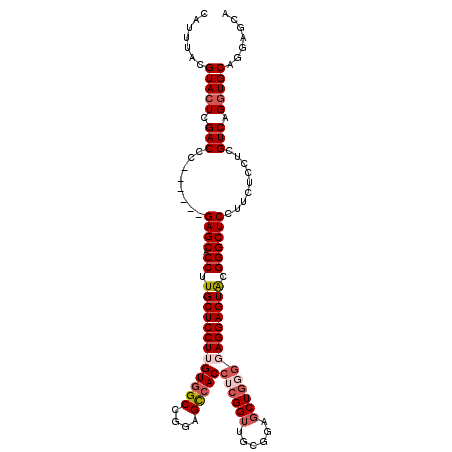
| Location | 20,102,791 – 20,102,898 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
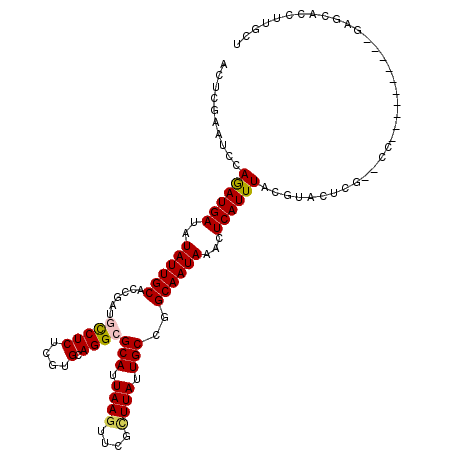
>3L_DroMel_CAF1 20102791 107 - 23771897 ACUCGAAUCCAGAUGAUAUAUUGCACCGAUGCCUCUCGUGCAGGCGCAUUAAGUUCGCUUAUUGCCGGCAAUAAACUCAUUUACGUACUCGACCC----------GAGCACCUUGCU .((((.....((((((..((((((......(((((....).))))(((.((((....)))).)))..))))))...)))))).((....))...)----------)))......... ( -26.50) >DroPse_CAF1 16134 115 - 1 UUUCAAAUCCAAAUGAAAUAUUGCACCGAAGUCUCUCGUGAAGGGGCAUUAAGGGGGUUUAUUGCCAGCAAUAAACUCAUUUGCGUACCCC--CCAAAUCCAGGGGAGACUCUUGUC (((((........)))))........(((((((((((.((..((((.((((((.((((((((((....)))))))))).)))).)).))))--.......)).)))))))).))).. ( -32.90) >DroSim_CAF1 17541 107 - 1 ACUCGAAUCCAGAUGAUAUAUUGCACCGAUGCCUCUCGUGCAGGCGCAUUAAGUUCGCUUACUGCCGGCAAUAAACUCAUUUACGUACUCGACCC----------GAGCACCUUGCU .((((.....((((((..((((((......(((((....).))))(((.((((....)))).)))..))))))...)))))).((....))...)----------)))......... ( -26.00) >DroYak_CAF1 17349 113 - 1 ACUCGAAUCCAGAUGAUAUAUUGCAACGAUGCCUCUCGUGCAGGCGCAUUAAGUUCGCUUAUUGCCGGCAAUAAACUCAUUUACGUACUCGACCCGUACUC----GAGCACCUUGCU .(((((....((((((..((((((......(((((....).))))(((.((((....)))).)))..))))))...))))))..((((.......))))))----)))......... ( -30.70) >DroAna_CAF1 15921 95 - 1 ACUCGAAUCCAGAUGAUAUAUUGCAUCGAUGCCUCUCGUGCAGGCGCAUUAACUCUGCUUAUUGCCGGCAAUAAACUCAUUUACGUAG----------------------CUUUCAU ..........((((((..((((((..(((((((((....).))))(((.......)))..))))...))))))...))))))......----------------------....... ( -18.40) >DroPer_CAF1 15999 115 - 1 UUUCAAAUCCAAAUGAAAUAUUGCACCGAAGUCUCUCGUGAAGGGGCAUUAAGGGGGUUUAUUGCCAGCAAUAAACUCAUUUGCGUACCCC--CCAAAUCCAGGGGAGACUCUUGUC (((((........)))))........(((((((((((.((..((((.((((((.((((((((((....)))))))))).)))).)).))))--.......)).)))))))).))).. ( -32.90) >consensus ACUCGAAUCCAGAUGAUAUAUUGCACCGAUGCCUCUCGUGCAGGCGCAUUAAGUUCGCUUAUUGCCGGCAAUAAACUCAUUUACGUACUCG__CC__________GAGCACCUUGCU ..........((((((..((((((......(((((....).))))(((.((((....)))).)))..))))))...))))))................................... (-17.64 = -17.53 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:31 2006