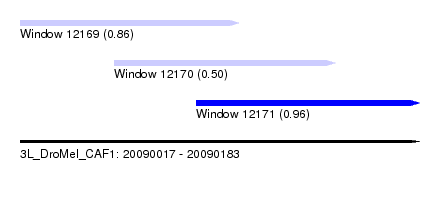
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,090,017 – 20,090,183 |
| Length | 166 |
| Max. P | 0.957382 |

| Location | 20,090,017 – 20,090,108 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -20.23 |
| Energy contribution | -23.47 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20090017 91 + 23771897 CUCUUUGAUGGCCCCUCCGAAGUGUGAAGAUUGCAUCUGGAGGCCGCGUGGGAAUGGGAUGUAGGUGAUUCUGCAUCCCAUCUG-------CCCACAA ......(.(((((..((((..(((..(...)..))).))))))))))(((((.((((((((((((....))))))))))))...-------))))).. ( -41.60) >DroSec_CAF1 4524 72 + 1 CUCUUUGAUGGCC-CUCCGAAGUGUGAAGAUUGCAUCUGGAGGUCGCGUGGAAAUGGGA---------UUCUGCAA----------------CCACAA ....(((.(((.(-(((((..(((..(...)..))).))))))....(..(((......---------)))..)..----------------)))))) ( -18.20) >DroSim_CAF1 4581 90 + 1 CUCUUUGAUGGCC-CUCCGAAGUGUGAAGAUUGCAUCUGGAGGUCGCGUGGAAAUGGGAUGUAGUUGAUUCUGCAUCCCAUCCG-------CCCACAA ....(((.(((.(-(((((..(((..(...)..))).))))))....(((((..((((((((((......))))))))))))))-------))))))) ( -36.30) >DroEre_CAF1 4507 96 + 1 CUCUUUGAUGGCC-CUCCGAAGUGUGAAGAUUGCAUCUGGAGG-CGCGUGGGCAUGGGUUGUUGCUGAUUCUGUAACCCAUCCGCAUCCAGCCCACAA ..........(((-(((((..(((..(...)..))).))))))-.))((((((.(((((.(((((.......)))))........))))))))))).. ( -34.00) >consensus CUCUUUGAUGGCC_CUCCGAAGUGUGAAGAUUGCAUCUGGAGGUCGCGUGGAAAUGGGAUGUAG_UGAUUCUGCAACCCAUCCG_______CCCACAA ......(.(((((..((((..(((..(...)..))).))))))))))(((((.(((((((((((......)))))))))))..........))))).. (-20.23 = -23.47 + 3.25)



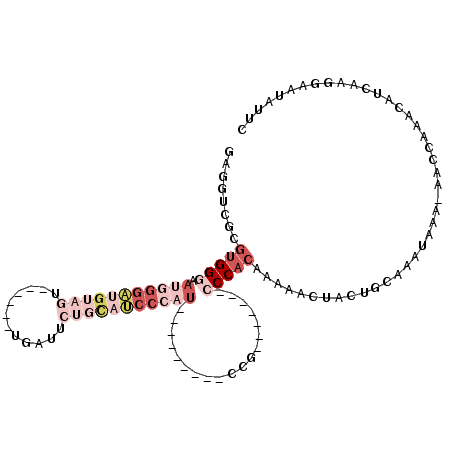
| Location | 20,090,056 – 20,090,148 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.28 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -5.63 |
| Energy contribution | -9.43 |
| Covariance contribution | 3.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.25 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20090056 92 + 23771897 GAGGCCGCGUGGGAAUGGGAUGUAGG------UGAUUCUGCAUCCCAU---------CUG-------CCCACAAAAACUACUGCAAAUAAAAAACCAAACAUCAAGGAAUAUUC ........(((((.((((((((((((------....))))))))))))---------...-------))))).......................................... ( -28.60) >DroSec_CAF1 4562 73 + 1 GAGGUCGCGUGGAAAUGGGA---------------UUCUGCAA-------------------------CCACAAAAACUACUGCAAAUAAA-AACCAAACAUCAAGGAAUAUUC ..((((.(((....))).))---------------...((((.-------------------------.............))))......-..)).................. ( -7.84) >DroSim_CAF1 4619 91 + 1 GAGGUCGCGUGGAAAUGGGAUGUAGU------UGAUUCUGCAUCCCAU---------CCG-------CCCACAAAAACUACUGCAAAUAAA-AACCAAACAUCAAGGAAUAUUC ...((.(.(((((..((((((((((.------.....)))))))))))---------)))-------)).))...................-...................... ( -24.10) >DroEre_CAF1 4545 96 + 1 GAGG-CGCGUGGGCAUGGGUUGUUGC------UGAUUCUGUAACCCAU---------CCGCAUCCAGCCCACAAAAACUACUGCGAAUAAA--AGCAAACAUCAAGAAAUAUUC ((..-...((((((.(((((.(((((------.......)))))....---------....))))))))))).........(((.......--.)))....))........... ( -22.10) >DroAna_CAF1 4450 108 + 1 GGGGGCG--AGGG-ACAGGACGUACUCACCCAGGACCCUUUAUCCCCUGAGCCCUCCCCCCAGCCUGCCCACAGAAACUACUG-AAAUAAA--AGCAAACAUCAAGAAAUAUUU (((((.(--((((-.((((..((....))...(((.......)))))))..)))))))))).((.......(((......)))-.......--.)).................. ( -28.86) >consensus GAGGUCGCGUGGGAAUGGGAUGUAGU______UGAUUCUGCAUCCCAU_________CCG_______CCCACAAAAACUACUGCAAAUAAA_AACCAAACAUCAAGGAAUAUUC ........(((((.(((((((((((............)))))))))))...................))))).......................................... ( -5.63 = -9.43 + 3.80)


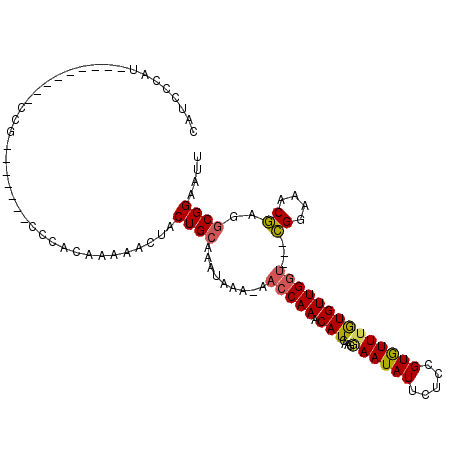
| Location | 20,090,090 – 20,090,183 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -11.85 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20090090 93 + 23771897 CAUCCCAU---------CUG-------CCCACAAAAACUACUGCAAAUAAAAAACCAAACAUCAAGGAAUAUUCUUCGUGUUUGUGUUGGU---CGGAAACGAGGCGGAAUU .......(---------(((-------((.......................((((((((((.(((((...))))).))))))).)))..(---((....)))))))))... ( -25.30) >DroSec_CAF1 4587 83 + 1 CAA-------------------------CCACAAAAACUACUGCAAAUAAA-AACCAAACAUCAAGGAAUAUUCUCCGUGUUUGUGUUGGU---CGGAAACGAGGCGGAAUU ...-------------------------............((((.......-((((((((((...(((......)))))))))).)))..(---((....))).)))).... ( -20.50) >DroSim_CAF1 4653 92 + 1 CAUCCCAU---------CCG-------CCCACAAAAACUACUGCAAAUAAA-AACCAAACAUCAAGGAAUAUUCUCCGUGUUGGUGUUGGU---CGGAAACGAGGCGGAAUU .......(---------(((-------((......................-..((((.(((((((((......)))...))))))))))(---((....)))))))))... ( -27.80) >DroAna_CAF1 4487 109 + 1 UAUCCCCUGAGCCCUCCCCCCAGCCUGCCCACAGAAACUACUG-AAAUAAA--AGCAAACAUCAAGAAAUAUUUUCCGUAUUUAUGUUGGCGGCGGGCAACCAGCCGGAAUU ..(((...((....))......(((((((..(((......)))-.......--.((.(((((....((((((.....))))))))))).)))))))))........)))... ( -24.40) >consensus CAUCCCAU_________CCG_______CCCACAAAAACUACUGCAAAUAAA_AACCAAACAUCAAGGAAUAUUCUCCGUGUUUGUGUUGGU___CGGAAACGAGGCGGAAUU ........................................((((.........(((((.(((....((((((.....))))))))))))))...((....))..)))).... (-11.85 = -11.98 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:24 2006