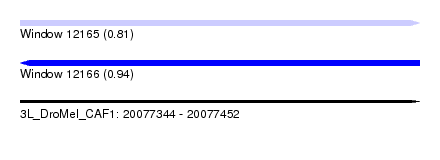
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,077,344 – 20,077,452 |
| Length | 108 |
| Max. P | 0.938773 |

| Location | 20,077,344 – 20,077,452 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -22.61 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
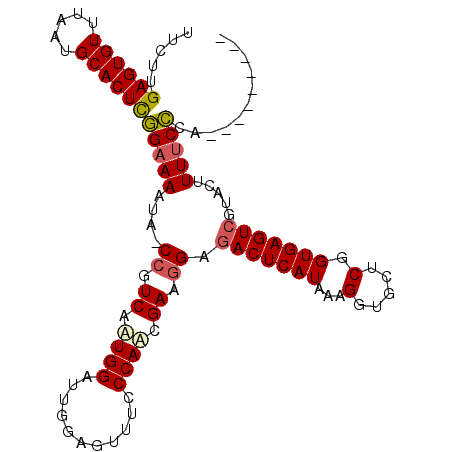
>3L_DroMel_CAF1 20077344 108 + 23771897 UUCUUGAGUGUUUAAUGCACUCGGAAAAUG-CCGUCAAUGGAUUGGAGUUUUCCAACGAAGGAAACUCAUAAAGGUGCUCGGUGAGUCGUACUUUUCCCAAGGUACUUU (((((((((((.....))))))(((((((.-(((((....)).))).))))))).....)))))((((((..((...))..)))))).(((((((....)))))))... ( -32.10) >DroSec_CAF1 15203 99 + 1 UUCUUGAGUGUUUAAUGCACUCGGAAAAUA-CCGUCAUUGGAUUGGAGUUUCCCAACGAAGGAGACUCAUAAAGGUGCUCGGUGAGUCGUACUUUUCCCA--------- .....((((((.....))))))((((((((-(((.((((......((((((((.......)))))))).....))))..)))))........))))))..--------- ( -31.40) >DroSim_CAF1 13769 99 + 1 UUCUUGAGUGUUUAAUGCACUCGGAAAAUA-CCGUCAAUGGAUUGGAGUUUCCCAACGAAGGAGACUCAUAAAGGUGCUCGGUGAGUCGAACUUUUCCCA--------- .....((((((.....))))))((((((((-(((((....)))..((((((((.......)))))))).....)))).((((....))))..))))))..--------- ( -29.50) >DroYak_CAF1 16896 99 + 1 GUCUUGAGUGUUUAAUGCACUUAGAAAAUAACCGUCAAUGGCUUGAAGUUUCCCAUCGA-CGAGACUCAUAAAGGUGCACGGUGAGUCGUACAUUACUCA--------- .(((.((((((.....))))))))).......((((.((((...........)))).))-)).(((((((...(.....).)))))))............--------- ( -25.60) >consensus UUCUUGAGUGUUUAAUGCACUCGGAAAAUA_CCGUCAAUGGAUUGGAGUUUCCCAACGAAGGAGACUCAUAAAGGUGCUCGGUGAGUCGUACUUUUCCCA_________ .....((((((.....))))))(((((....((.((.((((...........)))).)).)).(((((((...(.....).))))))).....)))))........... (-22.61 = -23.18 + 0.56)


| Location | 20,077,344 – 20,077,452 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -21.01 |
| Energy contribution | -22.07 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
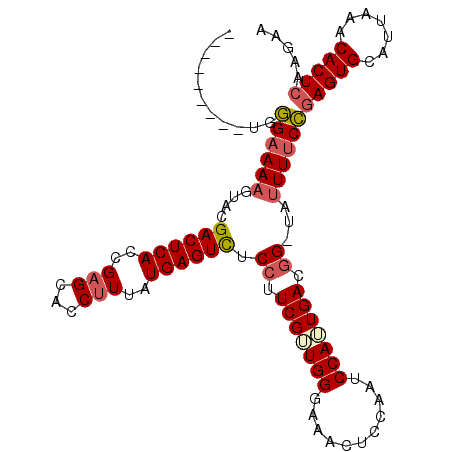
>3L_DroMel_CAF1 20077344 108 - 23771897 AAAGUACCUUGGGAAAAGUACGACUCACCGAGCACCUUUAUGAGUUUCCUUCGUUGGAAAACUCCAAUCCAUUGACGG-CAUUUUCCGAGUGCAUUAAACACUCAAGAA ...((((.((((((((.....((((((..(((...)))..)))))).((.(((.((((.........)))).))).))-..))))))))))))................ ( -24.20) >DroSec_CAF1 15203 99 - 1 ---------UGGGAAAAGUACGACUCACCGAGCACCUUUAUGAGUCUCCUUCGUUGGGAAACUCCAAUCCAAUGACGG-UAUUUUCCGAGUGCAUUAAACACUCAAGAA ---------..((((((....((((((..(((...)))..)))))).((.((((((((.........)))))))).))-..))))))(((((.......)))))..... ( -29.30) >DroSim_CAF1 13769 99 - 1 ---------UGGGAAAAGUUCGACUCACCGAGCACCUUUAUGAGUCUCCUUCGUUGGGAAACUCCAAUCCAUUGACGG-UAUUUUCCGAGUGCAUUAAACACUCAAGAA ---------..(((((((((((......)))))(((.....((((.((((.....)))).))))(((....)))..))-).))))))(((((.......)))))..... ( -28.80) >DroYak_CAF1 16896 99 - 1 ---------UGAGUAAUGUACGACUCACCGUGCACCUUUAUGAGUCUCG-UCGAUGGGAAACUUCAAGCCAUUGACGGUUAUUUUCUAAGUGCAUUAAACACUCAAGAC ---------((((((((((((((((((.............))))))(((-(((((((...........))))))))))...........)))))))....))))).... ( -30.32) >consensus _________UGGGAAAAGUACGACUCACCGAGCACCUUUAUGAGUCUCCUUCGUUGGGAAACUCCAAUCCAUUGACGG_UAUUUUCCGAGUGCAUUAAACACUCAAGAA ...........((((((....((((((..(((...)))..)))))).((.(((((((...........))))))).))...))))))(((((.......)))))..... (-21.01 = -22.07 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:20 2006