| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,035,046 – 20,035,226 |
| Length | 180 |
| Max. P | 0.880303 |

| Location | 20,035,046 – 20,035,136 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20035046 90 - 23771897 AAUGGGCGGAAAU------------GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUAAUCGGUGAGUUGCGGAAAAUAG (((..((.((...------------.(((((.(((((..((((.((((((.....))))))..)))).))))).)))))..)).))..)))........... ( -29.50) >DroSec_CAF1 2182 90 - 1 AAUGGGUGGAAAU------------GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUAAUCGGUGAGUUGUGGGAAAUAG .(..(.(.(....------------.(((((.(((((..((((.((((((.....))))))..)))).))))).)))))......).).)..)......... ( -26.60) >DroSim_CAF1 2181 90 - 1 AAUGGGUGGAAAU------------GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUAAUCGGUGAGUUGUGGGAAAUGG .(..(.(.(....------------.(((((.(((((..((((.((((((.....))))))..)))).))))).)))))......).).)..)......... ( -26.60) >DroEre_CAF1 6205 96 - 1 AAUGGGCAGGAGCGGCGGAAUCUUAGGAAGUGGGCGUGGCAGCUACGGCGAUACCCGCUGUCUGCUGGACGCUGACUAUAAUAGGUGAGU-----GAAA-AC ..(.(((((..(((((((..((....)).((.(.(((((...))))).).))..)))))))))))).).((((.(((......))).)))-----)...-.. ( -32.90) >DroYak_CAF1 2119 101 - 1 AAUGGGCAGGAAUGGCGGAAGCUUAGGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUCUGCUGGAUGGCGACUUCAAUAGGUGAGUUGAGGAAAA-AC ..(.((((((...(((....))).....((((((..(.((.......)).)..)))))).)))))).).......(((((((......)))))))....-.. ( -31.20) >consensus AAUGGGCGGAAAU____________GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUAAUCGGUGAGUUGUGGGAAAUAG ..........................(((((.(((((..((((.((((((.....))))))..)))).))))).)))))....................... (-24.94 = -24.70 + -0.24)


| Location | 20,035,068 – 20,035,176 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -46.18 |
| Consensus MFE | -29.86 |
| Energy contribution | -32.06 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
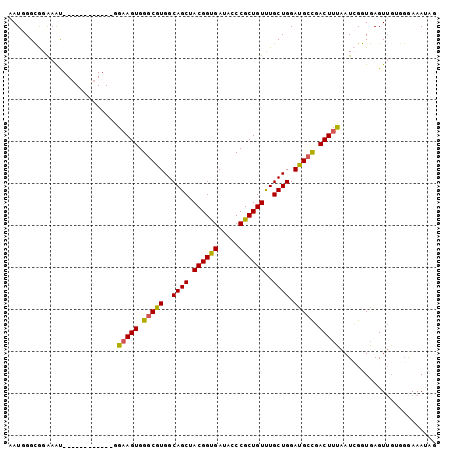
>3L_DroMel_CAF1 20035068 108 + 23771897 UAAAGUCGGCAUCCAGCAAACAGCGGGUAUCACCGUAGCUGCCACGCCCACUUCC------------AUUUCCGCCCAUUCUGUCGCCGCCGUGGGCGUGGCUGCCGCUGGAGCGGAAGA .....((.((.((((((.....((((......)))).((.(((((((((((....------------........................))))))))))).)).)))))))).))... ( -47.89) >DroSec_CAF1 2204 93 + 1 UAAAGUCGGCAUCCAGCAAACAGCGGGUAUCACCGUAGCUGCCACGCCCACUUCC------------AUUUCCACCCAUUCC---------------GUGGCUGCCGCUGGAGCGGAAGA .....((.((.((((((.....((((......)))).((.((((((.........------------..............)---------------))))).)).)))))))).))... ( -33.10) >DroSim_CAF1 2203 108 + 1 UAAAGUCGGCAUCCAGCAAACAGCGGGUAUCACCGUAGCUGCCACGCCCACUUCC------------AUUUCCACCCAUUCCGGCACCGCCGUGCGCGUGGCUGCCGCUGGAGCGGAAGA .....((.((.((((((.....((((......)))).((.(((((((.(((....------------...............(((...)))))).))))))).)).)))))))).))... ( -42.40) >DroEre_CAF1 6221 120 + 1 UAUAGUCAGCGUCCAGCAGACAGCGGGUAUCGCCGUAGCUGCCACGCCCACUUCCUAAGAUUCCGCCGCUCCUGCCCAUUGCGGCGCCGCCGUGGGCGUGGCUGGCGCUGGAGCGGAAGA .....((.((.((((((.....((((......)))).((((((((((((((.((....))...((((((...........)))))).....))))))))))).))))))))))).))... ( -58.30) >DroYak_CAF1 2140 117 + 1 UGAAGUCGCCAUCCAGCAGACAGCGGGUAUCACCGUAGCUGCCACGCCCACUUCCUAAGCUUCCGCCAUUCCUGCCCAUUCCGGCGCCGCCG---GAGUGGCUGGCGCUGGAGCGGAAGA .(((((.((......((((.(.((((......)))).)))))...))..))))).....(((((((...(((.((((((((((((...))))---)))))).....)).)))))))))). ( -49.20) >consensus UAAAGUCGGCAUCCAGCAAACAGCGGGUAUCACCGUAGCUGCCACGCCCACUUCC____________AUUUCCGCCCAUUCCGGCGCCGCCGUG_GCGUGGCUGCCGCUGGAGCGGAAGA .....((.((.((((((.....((((......)))).((.(((((((.(((........................................))).))))))).)).)))))))).))... (-29.86 = -32.06 + 2.20)

| Location | 20,035,068 – 20,035,176 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -50.18 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.42 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
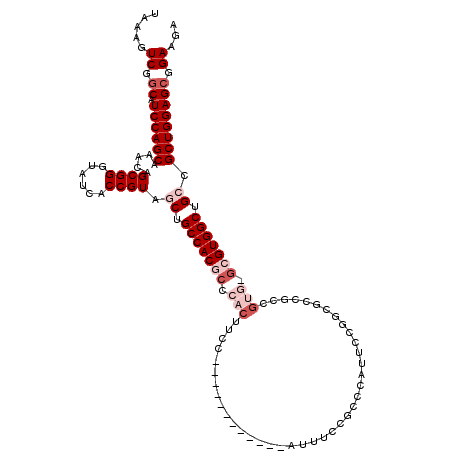
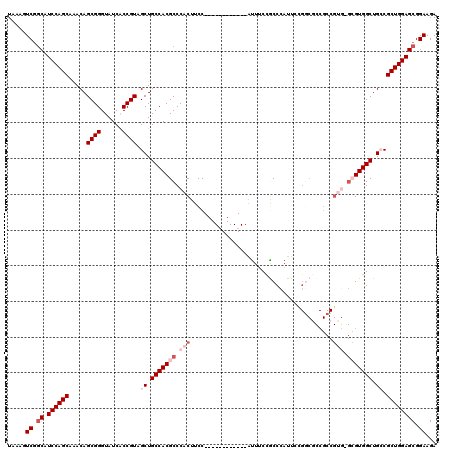
>3L_DroMel_CAF1 20035068 108 - 23771897 UCUUCCGCUCCAGCGGCAGCCACGCCCACGGCGGCGACAGAAUGGGCGGAAAU------------GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUA ...((.(((((((((((.(((((((((((..(.((..(.....).)).)....------------....))))))))))).)))((((((.....))))))..)))))).)).))..... ( -49.10) >DroSec_CAF1 2204 93 - 1 UCUUCCGCUCCAGCGGCAGCCAC---------------GGAAUGGGUGGAAAU------------GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUA ..((((((.(((.((.......)---------------)...)))))))))..------------.(((((.(((((..((((.((((((.....))))))..)))).))))).))))). ( -38.10) >DroSim_CAF1 2203 108 - 1 UCUUCCGCUCCAGCGGCAGCCACGCGCACGGCGGUGCCGGAAUGGGUGGAAAU------------GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUA ..((((((.(((.((((((((........)))..)))))...)))))))))..------------.(((((.(((((..((((.((((((.....))))))..)))).))))).))))). ( -49.70) >DroEre_CAF1 6221 120 - 1 UCUUCCGCUCCAGCGCCAGCCACGCCCACGGCGGCGCCGCAAUGGGCAGGAGCGGCGGAAUCUUAGGAAGUGGGCGUGGCAGCUACGGCGAUACCCGCUGUCUGCUGGACGCUGACUAUA (((.(((((((.((....))...(((((..(((....)))..))))).))))))).))).........(((.((((((((((..((((((.....)))))))))))..))))).)))... ( -60.60) >DroYak_CAF1 2140 117 - 1 UCUUCCGCUCCAGCGCCAGCCACUC---CGGCGGCGCCGGAAUGGGCAGGAAUGGCGGAAGCUUAGGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUCUGCUGGAUGGCGACUUCA .((((((((((.(((((.(((....---.)))))))).))).....((....))))))))).....(((((.(.((((((((..((((((.....)))))))))))..))).).))))). ( -53.40) >consensus UCUUCCGCUCCAGCGGCAGCCACGC_CACGGCGGCGCCGGAAUGGGCGGAAAU____________GGAAGUGGGCGUGGCAGCUACGGUGAUACCCGCUGUUUGCUGGAUGCCGACUUUA ..((((((.(((......(((.(.......).))).......)))))))))...............(((((.(((((..((((.((((((.....))))))..)))).))))).))))). (-37.06 = -37.42 + 0.36)

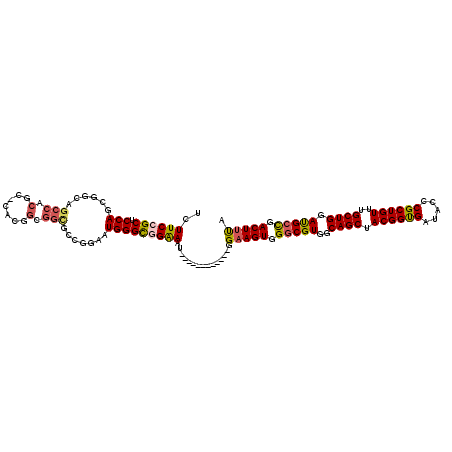

| Location | 20,035,136 – 20,035,226 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -35.88 |
| Energy contribution | -37.38 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20035136 90 + 23771897 CUGUCGCCGCCGUGGGCGUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCGAGGCGACCU ..((((((..((..(((((..(.(((((.(((((....((((((.....))))))...)))))))))).)..)))))..)).)))))).. ( -54.40) >DroSec_CAF1 2272 75 + 1 CC---------------GUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCGAGGCGACCU ..---------------((.((((((((.(((((....((((((.....))))))...))))))))))(((.......))).))).)).. ( -37.90) >DroSim_CAF1 2271 90 + 1 CCGGCACCGCCGUGCGCGUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCAAGGCGACCU ..((...(((((((.((((..(.(((((.(((((....((((((.....))))))...)))))))))).)..)))))))...)))).)). ( -46.20) >DroYak_CAF1 2220 87 + 1 CCGGCGCCGCCG---GAGUGGCUGGCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUACCAUCGAGGCGACCU (((((((((((.---....))).)))))))).(((((.((((((.....)))))).....))))).(((..(((...)))..)))..... ( -45.60) >consensus CCGGCGCCGCCG___GCGUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCGAGGCGACCU .......((((......(((((.(((((.(((((....((((((.....))))))...))))))))))......)))))...)))).... (-35.88 = -37.38 + 1.50)


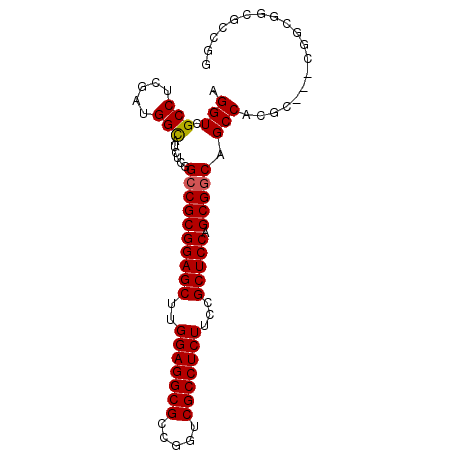
| Location | 20,035,136 – 20,035,226 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -38.06 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20035136 90 - 23771897 AGGUCGCCUCGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCACGCCCACGGCGGCGACAG ..((((((..(.((((......((((((((((..(((((((.....)))))))...))))).))))).)))))(((...))))))))).. ( -51.50) >DroSec_CAF1 2272 75 - 1 AGGUCGCCUCGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCAC---------------GG .(((.(((.....)))......((((((((((..(((((((.....)))))))...))))).))))).)))..---------------.. ( -40.40) >DroSim_CAF1 2271 90 - 1 AGGUCGCCUUGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCACGCGCACGGCGGUGCCGG .(((.(((.....)))......((((((((((..(((((((.....)))))))...))))).))))).))).((.((((....)))))). ( -47.80) >DroYak_CAF1 2220 87 - 1 AGGUCGCCUCGAUGGUAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGCCAGCCACUC---CGGCGGCGCCGG ((((.(((.....)))))))((((.(((((((..(((((((.....)))))))...))))).))(((.(((....---.)))))))))). ( -44.40) >consensus AGGUCGCCUCGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCACGC___CGGCGGCGCCGG .(((.(((.....)))......((((((((((..(((((((.....)))))))...))))).))))).)))................... (-38.06 = -38.12 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:09 2006