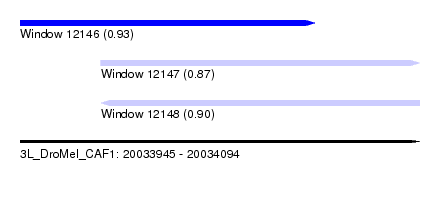
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,033,945 – 20,034,094 |
| Length | 149 |
| Max. P | 0.927640 |

| Location | 20,033,945 – 20,034,055 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.59 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20033945 110 + 23771897 CAAAUUGAGGUCACACCAUCGCCACCACCACACCAUCAGUUCCACGCACACUAUUAUCUAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUGAAGGAAGU ......((((.....)).))..............(((.(((((((((................)))))))))))).(((((((.(((......)))...))).))))... ( -23.59) >DroSec_CAF1 1086 102 + 1 CAAAUUGAGGUCACACCAUCGCCAUCA-----CCAUCAGUUCCACACACACUA---UCCAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUAAAGGAAGU .....((((((.........))).)))-----((....(((((((((......---.......)))))))))(((((....)))))..................)).... ( -20.92) >DroSim_CAF1 1082 105 + 1 CAAAUUGAGGUCACACCAUCGCCAUCA-----CCAUCAGUUCCACACACACUAUCAUCCAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUGAAGGAAGU .....((((((.........))).)))-----..(((.(((((((((................)))))))))))).(((((((.(((......)))...))).))))... ( -24.49) >consensus CAAAUUGAGGUCACACCAUCGCCAUCA_____CCAUCAGUUCCACACACACUAU_AUCCAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUGAAGGAAGU ......((((.....)).))..............(((.(((((((((................)))))))))))).(((((((.(((......)))...))).))))... (-22.48 = -22.59 + 0.11)



| Location | 20,033,975 – 20,034,094 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -26.88 |
| Energy contribution | -27.69 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20033975 119 + 23771897 CACCAUCAGUUCCACGCACACUAUUAUCUAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUGAAGGAAGUAUGUAGAACCGGAGGUGCGAGGGCUAACUACCCACUUCC- ....(((.(((((((((................)))))))))))).(((((((.(((......)))...))).)))).............(((((((.(.((.....)).).)))))))- ( -31.89) >DroSec_CAF1 1113 114 + 1 --CCAUCAGUUCCACACACACUA---UCCAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUAAAGGAAGUAUGCAGAACCGGAGGUGCGAGGGCUAACUACCCACUUCC- --..(((.(((((((((......---.......)))))))))))).((((.((.(((......)))...))..)))).............(((((((.(.((.....)).).)))))))- ( -28.72) >DroSim_CAF1 1109 117 + 1 --CCAUCAGUUCCACACACACUAUCAUCCAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUGAAGGAAGUAUGUAGAGCCGGAGGUGCGAGGGCUAACUACCCACUUCC- --..(((.(((((((((................))))))))))))....((((.(((......)))...)))).((((((..((((((((...........))))..))))..))))))- ( -32.89) >DroYak_CAF1 1004 103 + 1 --CCAUCAGUUCCACACACU-------------UUGUGGAACCAUAUCCUCG--UCUAGACAAAGACAAGUGAACGAAGUAUGCAAAACCGGAGGUGCGAGGGCUAACUACCCACUUCCA --......((((((((....-------------.))))))))(((((..(((--(((......))))..(....))).))))).......(((((((.(.((.....)).).))))))). ( -29.50) >consensus __CCAUCAGUUCCACACACACUA___UCCAUCAGUGUGGAACGAUAUCCUCGUGUCUAGACAAAGAAAAAUGAAGGAAGUAUGCAGAACCGGAGGUGCGAGGGCUAACUACCCACUUCC_ ........(((((((((................)))))))))....(((((((.(((......)))...))).)))).............(((((((.(.((.....)).).))))))). (-26.88 = -27.69 + 0.81)


| Location | 20,033,975 – 20,034,094 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -30.42 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20033975 119 - 23771897 -GGAAGUGGGUAGUUAGCCCUCGCACCUCCGGUUCUACAUACUUCCUUCAUUUUUCUUUGUCUAGACACGAGGAUAUCGUUCCACACUGAUAGAUAAUAGUGUGCGUGGAACUGAUGGUG -....(.((((.....)))).).((((..(((((((((................(((((((......)))))))........(((((((........))))))).)))))))))..)))) ( -37.00) >DroSec_CAF1 1113 114 - 1 -GGAAGUGGGUAGUUAGCCCUCGCACCUCCGGUUCUGCAUACUUCCUUUAUUUUUCUUUGUCUAGACACGAGGAUAUCGUUCCACACUGAUGGA---UAGUGUGUGUGGAACUGAUGG-- -(((((((.((((..((((...........)))))))).)))))))........(((((((......)))))))(((((((((((((.......---......))))))))).)))).-- ( -36.32) >DroSim_CAF1 1109 117 - 1 -GGAAGUGGGUAGUUAGCCCUCGCACCUCCGGCUCUACAUACUUCCUUCAUUUUUCUUUGUCUAGACACGAGGAUAUCGUUCCACACUGAUGGAUGAUAGUGUGUGUGGAACUGAUGG-- -(((((((.((((..((((...........)))))))).)))))))........(((((((......)))))))(((((((((((((..((........))..))))))))).)))).-- ( -38.60) >DroYak_CAF1 1004 103 - 1 UGGAAGUGGGUAGUUAGCCCUCGCACCUCCGGUUUUGCAUACUUCGUUCACUUGUCUUUGUCUAGA--CGAGGAUAUGGUUCCACAA-------------AGUGUGUGGAACUGAUGG-- .....(.((((.....)))).)...((..((((((..(((((((.((..((((((((((((....)--)))))))).)))...)).)-------------))))))..))))))..))-- ( -37.70) >consensus _GGAAGUGGGUAGUUAGCCCUCGCACCUCCGGUUCUACAUACUUCCUUCAUUUUUCUUUGUCUAGACACGAGGAUAUCGUUCCACACUGAUGGA___UAGUGUGUGUGGAACUGAUGG__ .(((((((.((((..((((...........)))))))).)))))))........(((((((......)))))))(((((((((((((................))))))))).))))... (-30.42 = -31.04 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:02 2006