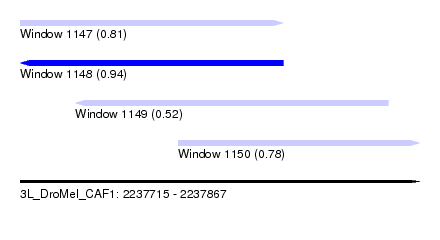
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,237,715 – 2,237,867 |
| Length | 152 |
| Max. P | 0.936725 |

| Location | 2,237,715 – 2,237,815 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2237715 100 + 23771897 UACAUGUUCCACCUGCCCGCAGAUUAAAAUAACCCCCAAGCACUUGCUGAUAACAUGCGACCAGAAGUUUAGGGAGCCAAUAGGCUUCCCAAAAACCCAC ..((((((....(((....)))................(((....)))...)))))).........((((.(((((((....)))))))...)))).... ( -21.50) >DroSec_CAF1 18639 100 + 1 UACAUGUUCCACCUGCCCUCAGAUUAAAAUAACCCCCAAGUACUUGCUGAUAACAUGCGACCAGAAGUUUAGGGAGCCAGUAGGCUUCCCAAAAACCCAC ..((((((....(((....)))................(((....)))...)))))).........((((.(((((((....)))))))...)))).... ( -17.40) >DroSim_CAF1 18714 100 + 1 UACAUGUUCCACCUGCCCGCAGAUUAAAAUAACCCCCAAGCAGUUGCUGAUAACAUGCGACCAAAAGUUUAGGGAGCCAGUAGGCUCCCCAAAAACCCAC ..((((((....(((....)))................(((....)))...)))))).........((((.(((((((....)))))))...)))).... ( -24.40) >consensus UACAUGUUCCACCUGCCCGCAGAUUAAAAUAACCCCCAAGCACUUGCUGAUAACAUGCGACCAGAAGUUUAGGGAGCCAGUAGGCUUCCCAAAAACCCAC ..((((((....(((....)))................(((....)))...)))))).........((((.(((((((....)))))))...)))).... (-21.54 = -21.10 + -0.44)

| Location | 2,237,715 – 2,237,815 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -32.23 |
| Energy contribution | -32.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2237715 100 - 23771897 GUGGGUUUUUGGGAAGCCUAUUGGCUCCCUAAACUUCUGGUCGCAUGUUAUCAGCAAGUGCUUGGGGGUUAUUUUAAUCUGCGGGCAGGUGGAACAUGUA ..(((..((((((.((((....)))).))))))..)))....(((((((.(((.(...((((((.((((((...)))))).))))))).)))))))))). ( -36.20) >DroSec_CAF1 18639 100 - 1 GUGGGUUUUUGGGAAGCCUACUGGCUCCCUAAACUUCUGGUCGCAUGUUAUCAGCAAGUACUUGGGGGUUAUUUUAAUCUGAGGGCAGGUGGAACAUGUA ..(((..((((((.((((....)))).))))))..)))....(((((((....((..(..((..((...........))..))..)..))..))))))). ( -29.60) >DroSim_CAF1 18714 100 - 1 GUGGGUUUUUGGGGAGCCUACUGGCUCCCUAAACUUUUGGUCGCAUGUUAUCAGCAACUGCUUGGGGGUUAUUUUAAUCUGCGGGCAGGUGGAACAUGUA .(..(..(((((((((((....)))))))))))..)..)...(((((((.(((....(((((((.((((((...)))))).))))))).)))))))))). ( -43.00) >consensus GUGGGUUUUUGGGAAGCCUACUGGCUCCCUAAACUUCUGGUCGCAUGUUAUCAGCAAGUGCUUGGGGGUUAUUUUAAUCUGCGGGCAGGUGGAACAUGUA ..((((((..(((.((((....))))))).))))))......(((((((.(((.(...((((((.((((((...)))))).))))))).)))))))))). (-32.23 = -32.90 + 0.67)


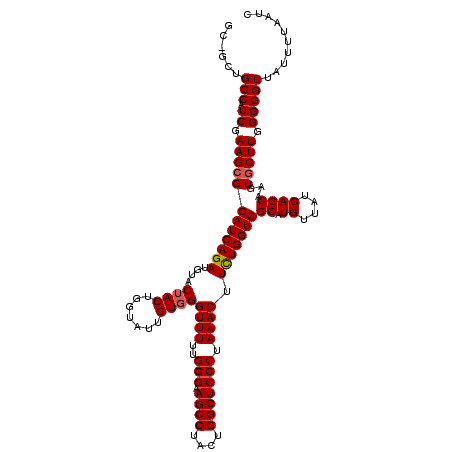
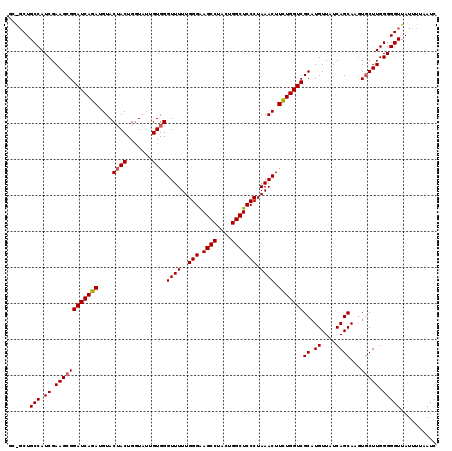
| Location | 2,237,736 – 2,237,855 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -33.82 |
| Energy contribution | -34.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2237736 119 - 23771897 GCAGCUGCCAUCGAAGCGGAUCAGAUGUACAACUGUUAAAGUGGGUUUUUGGGAAGCCUAUUGGCUCCCUAAACUUCUGGUCGCAUGUUAUCAGCAAGUGCUUGGGGGUUAUUUUAAUC ..((((.(((....(((((((((((......(((.....))).(((((..((((.(((....))))))).))))))))))))((.((....))))...))))))).))))......... ( -36.40) >DroSec_CAF1 18660 119 - 1 GCGGCUGCCAUCGAAGCGGAUCAGAUGUACUACUGGUAUUGUGGGUUUUUGGGAAGCCUACUGGCUCCCUAAACUUCUGGUCGCAUGUUAUCAGCAAGUACUUGGGGGUUAUUUUAAUC ..((((.(((.....(((.((((((....((((.......))))((((..((((.(((....))))))).)))).))))))))).(((.....)))......))).))))......... ( -36.20) >DroSim_CAF1 18735 115 - 1 ----CUGCCAUCGAAGCGGAUCAGAUGUACUACUGGUAUUGUGGGUUUUUGGGGAGCCUACUGGCUCCCUAAACUUUUGGUCGCAUGUUAUCAGCAACUGCUUGGGGGUUAUUUUAAUC ----..(((.((.((((((((((((.((.((((.......))))....((((((((((....)))))))))))).)))))))((.((....))))...))))).))))).......... ( -41.60) >consensus GC_GCUGCCAUCGAAGCGGAUCAGAUGUACUACUGGUAUUGUGGGUUUUUGGGAAGCCUACUGGCUCCCUAAACUUCUGGUCGCAUGUUAUCAGCAAGUGCUUGGGGGUUAUUUUAAUC ......(((.((.((((((((((((....((((.......))))((((..(((.((((....))))))).)))).)))))))((.((....))))...))))).))))).......... (-33.82 = -34.27 + 0.45)

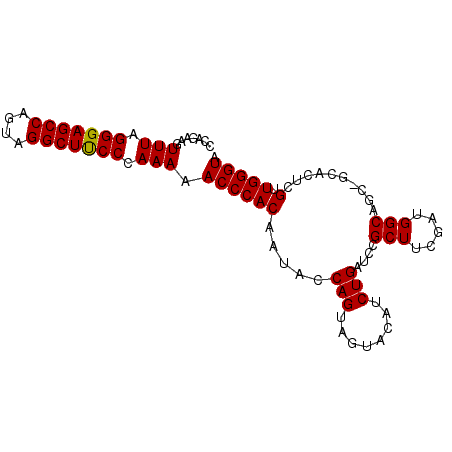
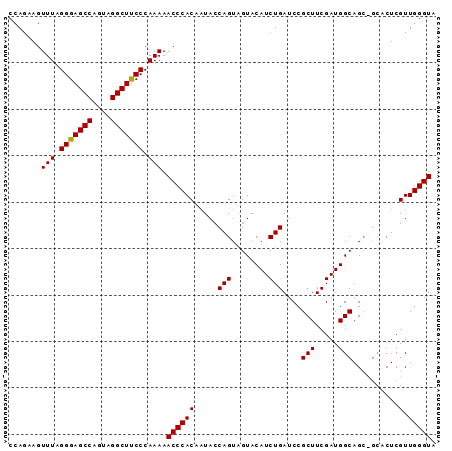
| Location | 2,237,775 – 2,237,867 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.94 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2237775 92 + 23771897 CCAGAAGUUUAGGGAGCCAAUAGGCUUCCCAAAAACCCACUUUAACAGUUGUACAUCUGAUCCGCUUCGAUGGCAGCUGCACUCGUUGGGUA .......(((.(((((((....))))))).))).(((((......(((((((.((((.((......))))))))))))).......))))). ( -31.32) >DroSec_CAF1 18699 92 + 1 CCAGAAGUUUAGGGAGCCAGUAGGCUUCCCAAAAACCCACAAUACCAGUAGUACAUCUGAUCCGCUUCGAUGGCAGCCGCACUCGUUGGGUA .......(((.(((((((....))))))).))).(((((....((.(((....((((.((......))))))((....))))).))))))). ( -25.20) >DroSim_CAF1 18774 87 + 1 CCAAAAGUUUAGGGAGCCAGUAGGCUCCCCAAAAACCCACAAUACCAGUAGUACAUCUGAUCCGCUUCGAUGGCAG-----CUCGUUGGGUA .......(((.(((((((....))))))).))).(((((....((.(((.((.((((.((......)))))))).)-----)).))))))). ( -28.80) >consensus CCAGAAGUUUAGGGAGCCAGUAGGCUUCCCAAAAACCCACAAUACCAGUAGUACAUCUGAUCCGCUUCGAUGGCAGC_GCACUCGUUGGGUA .......(((.(((((((....))))))).))).((((((.....(((........)))....(((.....)))..........).))))). (-24.49 = -24.27 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:10 2006