
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,029,447 – 20,029,594 |
| Length | 147 |
| Max. P | 0.991168 |

| Location | 20,029,447 – 20,029,554 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -34.37 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
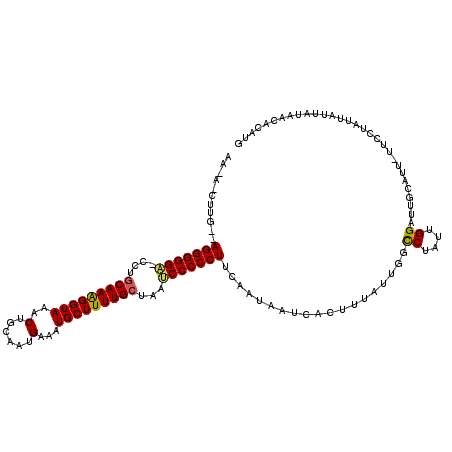
>3L_DroMel_CAF1 20029447 107 + 23771897 GCUCACAUGAGUGGCCAGAGGCG------------GAGACAACAACUUGCCAGGGGGC-CUUGCAAAGGUAAACUGCAAUGAAUUGCUUUUGCUAAUCCCCCUUCAAUAAUCACUUUUUU ((((....))))(((.((..(.(------------....)..)..)).)))((((((.-...((((((((((...........))))))))))....))))))................. ( -32.00) >DroSim_CAF1 16287 120 + 1 GCUCACACGAGUGGCCAGAGGCGGACGCGGAGGUGGAGGCGAGAUCUUGUUAGGGGGUACCAACAAAGGUAAACUGCAAUGAACUGCUUUUGCUAAGCCCCCUUCAAUAAUCACUUUAUU ((.(((....)))))..((((.((..((...((..(((((.((.((((((...((..((((......))))..)))))).)).)))))))..))..)).))))))............... ( -33.80) >DroYak_CAF1 22857 97 + 1 GCUCACACGAGUGCGCAGAGGCG------------CAGGUA----------AGGGGGA-UCUGCAAGGGUAAACUGCAAUGAAAUGCUUUUGCUAAUCCCCCUUAAAAAAUCACCUUAUU ((((....))))((((....)))------------)...((----------(((((((-(..(((((((((..(......)...)))))))))..))))))))))............... ( -37.30) >consensus GCUCACACGAGUGGCCAGAGGCG____________GAGGCAA_A_CUUG__AGGGGGA_CCUGCAAAGGUAAACUGCAAUGAAAUGCUUUUGCUAAUCCCCCUUCAAUAAUCACUUUAUU ((((....)))).......................................(((((((....(((((((((..(......)...)))))))))...)))))))................. (-22.44 = -22.67 + 0.23)



| Location | 20,029,475 – 20,029,594 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.32 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -19.89 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20029475 119 + 23771897 AACAACUUGCCAGGGGGC-CUUGCAAAGGUAAACUGCAAUGAAUUGCUUUUGCUAAUCCCCCUUCAAUAAUCACUUUUUUGGCCUAUUGGAUUGCCUUGUUCCUAUUAUUAUAACACAUG (((((...((..(((((.-...((((((((((...........))))))))))....)))))(((((((.(((......)))..)))))))..)).)))))................... ( -26.60) >DroSim_CAF1 16327 120 + 1 GAGAUCUUGUUAGGGGGUACCAACAAAGGUAAACUGCAAUGAACUGCUUUUGCUAAGCCCCCUUCAAUAAUCACUUUAUUGGCCUAUUGGAUUGCAUUAUUCCUAUUAUUAUAACACAUG .......(((((((((((.....((((((((.............))))))))....)))))).(((((((.....)))))))......(((.........)))........))))).... ( -24.72) >DroYak_CAF1 22885 100 + 1 A----------AGGGGGA-UCUGCAAGGGUAAACUGCAAUGAAAUGCUUUUGCUAAUCCCCCUUAAAAAAUCACCUUAUUGGUCCAUGGGAUUGCA---------UUAUUAUAACAGACG (----------(((((((-(..(((((((((..(......)...)))))))))..)))))))))....((((.((............))))))...---------............... ( -28.90) >consensus AA_A_CUUG__AGGGGGA_CCUGCAAAGGUAAACUGCAAUGAAAUGCUUUUGCUAAUCCCCCUUCAAUAAUCACUUUAUUGGCCUAUUGGAUUGCAUU_UUCCUAUUAUUAUAACACAUG ...........(((((((....(((((((((..(......)...)))))))))...)))))))...................((....)).............................. (-19.89 = -19.90 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:56 2006