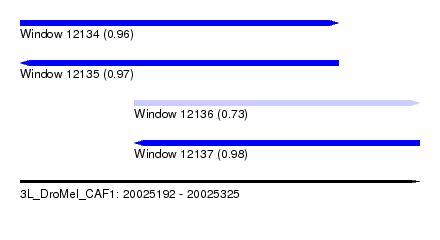
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,025,192 – 20,025,325 |
| Length | 133 |
| Max. P | 0.979548 |

| Location | 20,025,192 – 20,025,298 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20025192 106 + 23771897 UUUCCCUCUCAGCUGCUCAGUUGACGGUGAACUUCGCUGUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUGUGUGUACAUAUAUAUGUAUA .........(((((....)))))((((((.....)))))).....(((((.((((.(((((((((..(((.((....)).))).))))))))).)))).))))).. ( -26.70) >DroSec_CAF1 16903 102 + 1 UUUCCCUCUCAGCUGCUCAGUUGACGCUGAACUUCGCUGUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUUUGUGUACAUACU----UAUA ......((.((((.(.(((((....))))).)...)))).))..((((....))))((((((((((.(((......))).))))...)))))).....----.... ( -21.40) >DroSim_CAF1 11547 102 + 1 UUUCCCUCUCAGCUGCUCAGUUGACGCUGAACUUCGCUGUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUUUGUGUACCUACA----UAUA ......((.((((.(.(((((....))))).)...)))).))...........(((((((((((((.(((......))).))))...)))))))))..----.... ( -25.90) >DroEre_CAF1 16280 94 + 1 UUUCCCUCUCAGCUGCUCAGUUGACGCUGAACUUUGCUGUGUUUCUAUAUUUAUAGGUACAUACAUAUGUUUGCCUACAUAUGUUUUAUGUAC------------A .......(.((((.(.(((((....))))).)...)))).)...((((....))))((((((((((((((......))))))))...))))))------------. ( -24.50) >DroYak_CAF1 18582 102 + 1 UUUCCCUCUCAGUUGCUCAGUUGACGCUGAACUUUGCUGUGAUUCUAUAUUUAUAAGUUCAUACAUAUGUUUGUCUACAUUUGUUUUAUUUACUUACU----UAUA ......((.((((.(.(((((....))))).)...)))).))...........(((((....(((.((((......)))).))).......)))))..----.... ( -16.40) >consensus UUUCCCUCUCAGCUGCUCAGUUGACGCUGAACUUCGCUGUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUUUGUGUACAUACA____UAUA ......((.((((.(.(((((....))))).)...)))).))..............((((((((((.(((......))).))))...))))))............. (-16.58 = -17.18 + 0.60)

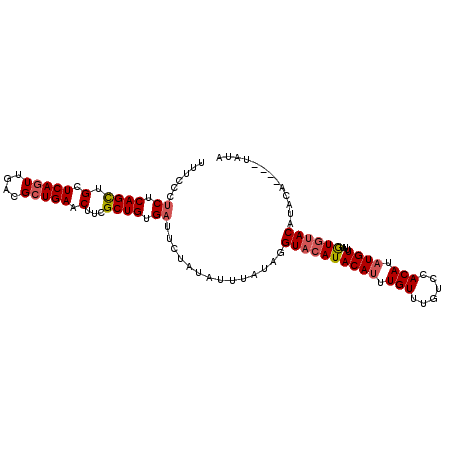
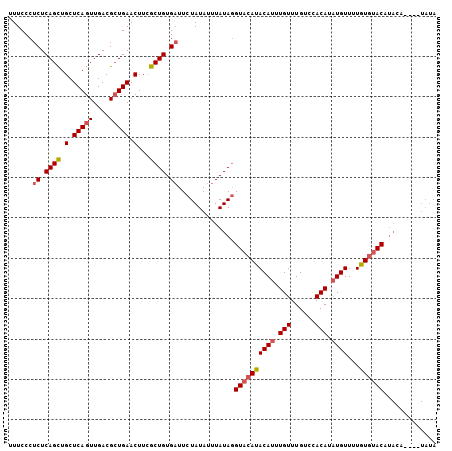
| Location | 20,025,192 – 20,025,298 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.98 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20025192 106 - 23771897 UAUACAUAUAUAUGUACACACAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCACAGCGAAGUUCACCGUCAACUGAGCAGCUGAGAGGGAAA ..(((((((((.(((...((((.....)))).....))).))))))))).((((....))))..((.((((...(((((........))))).)))).))...... ( -24.20) >DroSec_CAF1 16903 102 - 1 UAUA----AGUAUGUACACAAAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCACAGCGAAGUUCAGCGUCAACUGAGCAGCUGAGAGGGAAA ....----.(((.(((((....((((.(((......))).)))).))))).)))..........((.((((...((((((......)))))).)))).))...... ( -23.20) >DroSim_CAF1 11547 102 - 1 UAUA----UGUAGGUACACAAAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCACAGCGAAGUUCAGCGUCAACUGAGCAGCUGAGAGGGAAA (((.----((((((((((....((((.(((......))).)))).)))))))))).))).....((.((((...((((((......)))))).)))).))...... ( -31.00) >DroEre_CAF1 16280 94 - 1 U------------GUACAUAAAACAUAUGUAGGCAAACAUAUGUAUGUACCUAUAAAUAUAGAAACACAGCAAAGUUCAGCGUCAACUGAGCAGCUGAGAGGGAAA .------------((((((...((((((((......))))))))))))))((((....)))).....((((...((((((......)))))).))))......... ( -25.40) >DroYak_CAF1 18582 102 - 1 UAUA----AGUAAGUAAAUAAAACAAAUGUAGACAAACAUAUGUAUGAACUUAUAAAUAUAGAAUCACAGCAAAGUUCAGCGUCAACUGAGCAACUGAGAGGGAAA ....----.((((((..((...(((.((((......)))).)))))..))))))..........((.(((....((((((......))))))..))).))...... ( -15.90) >consensus UAUA____AGUAUGUACACAAAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCACAGCGAAGUUCAGCGUCAACUGAGCAGCUGAGAGGGAAA .............(((((....((((.(((......))).)))).)))))((((....))))..((.((((...((((((......)))))).)))).))...... (-16.94 = -17.98 + 1.04)

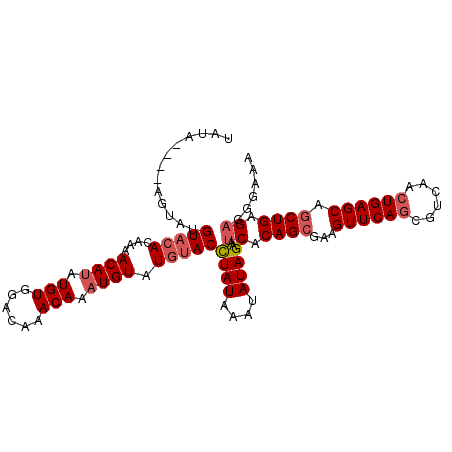
| Location | 20,025,230 – 20,025,325 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -10.71 |
| Energy contribution | -10.53 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20025230 95 + 23771897 GUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUGUGUGUACAUAUAUAUGUAUAUACUCGUAUUUUUCCUCAUAUAUUGAU ((((.........((..((((.((((((.(((.(((.(((((.....))))).))).))).)))))).))))..)).......))))........ ( -19.79) >DroSec_CAF1 16941 91 + 1 GUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUUUGUGUACAUACU----UAUAUACUCGUGUUUUUCCUCAUAUAUUGAU ((((..((((....))))((((((((((.(((......))).))))...)))))).....----...................))))........ ( -12.10) >DroSim_CAF1 11585 91 + 1 GUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUUUGUGUACCUACA----UAUAUACUCGUAUUUUUCCUCAUAUAUUGAU .......(((((...(((((((((((((.(((......))).))))...)))))))))..----.))))).............(((.....))). ( -17.00) >DroEre_CAF1 16318 83 + 1 GUGUUUCUAUAUUUAUAGGUACAUACAUAUGUUUGCCUACAUAUGUUUUAUGUAC------------AUACAAGUAUUUUUCCUCAUAUAUUGAA (((.....((((((.((.((((((((((((((......))))))))...))))))------------.)).)))))).......)))........ ( -16.70) >consensus GUGAUUCUAUAUUUAUAGGUACAUACAUUUGUUUGUCCACAUAUGUUUUGUGUACAUACA____UAUAUACUCGUAUUUUUCCUCAUAUAUUGAU (((...((((....))))((((((((((.(((......))).))))...)))))).............)))............(((.....))). (-10.71 = -10.53 + -0.19)


| Location | 20,025,230 – 20,025,325 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20025230 95 - 23771897 AUCAAUAUAUGAGGAAAAAUACGAGUAUAUACAUAUAUAUGUACACACAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCAC .....(((((.(((....(((....))).(((((((((.(((...((((.....)))).....))).))))))))))))....)))))....... ( -16.70) >DroSec_CAF1 16941 91 - 1 AUCAAUAUAUGAGGAAAAACACGAGUAUAUA----AGUAUGUACACAAAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCAC .....(((((.(((..........((((((.----...))))))(((..((((.(((......))).)))).))).)))....)))))....... ( -14.00) >DroSim_CAF1 11585 91 - 1 AUCAAUAUAUGAGGAAAAAUACGAGUAUAUA----UGUAGGUACACAAAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCAC ......................((.(((((.----((((((((((....((((.(((......))).)))).)))))))))).)))))...)).. ( -20.70) >DroEre_CAF1 16318 83 - 1 UUCAAUAUAUGAGGAAAAAUACUUGUAU------------GUACAUAAAACAUAUGUAGGCAAACAUAUGUAUGUACCUAUAAAUAUAGAAACAC (((...(((..((........))..)))------------((((((...((((((((......))))))))))))))...........))).... ( -17.80) >consensus AUCAAUAUAUGAGGAAAAAUACGAGUAUAUA____UGUAUGUACACAAAACAUAUGUGGACAAACAAAUGUAUGUACCUAUAAAUAUAGAAUCAC .....(((((.(((..........(((((((......)))))))(((..((((.(((......))).)))).))).)))....)))))....... (-12.73 = -12.97 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:52 2006