
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,024,122 – 20,024,232 |
| Length | 110 |
| Max. P | 0.657681 |

| Location | 20,024,122 – 20,024,232 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -19.58 |
| Energy contribution | -21.78 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20024122 110 - 23771897 AAGUAAGGAUGAGUCGUCUCAGGUAGAAAGUUGCCAUUGUUUAUGGUAAUCGGCUAAAGG-ACUAAAGAGUGAGGCUUCGCAGGGAG-AUGGGAUUCUGAAAAGCGGCCACU .........((.(((((((....(((...((((((((.....))))))))...)))..))-))...........((((..(((((..-......)))))..))))))))).. ( -25.70) >DroSec_CAF1 15874 110 - 1 AAAUAAGGAUGAGUCGUCUCAGGUAGAAAGUUGCCAUUGUUUAUGGUAAUCGGCUAAAGG-ACUAAAGAGUGAGGCUGCGCAGGGAG-AUGGGACUCUGAAAAGCGGCCACU ...............((((....(((...((((((((.....))))))))...)))..))-))..........((((((.(((((..-......)))))....))))))... ( -31.90) >DroSim_CAF1 10528 110 - 1 AAAUAAGGAUGAGUCGUCUCAGGUAGAAAGUUGCCAUUGUUUAUGGUAAUCGGCUAAAGG-ACUAAAGAGUGAGGCUGCGCAGGGAG-AUGGGACUCUGAAAGGCGGCCACU ...............((((....(((...((((((((.....))))))))...)))..))-))..........((((((.(((((..-......)))))....))))))... ( -30.80) >DroEre_CAF1 15346 112 - 1 AAAUAAGGAUGAGUCGUCUCAGGUAGAAAGUUGCCAUUGUUUAUGGUAAUCGGCUAAAGGGCCGAAAGAGUGGGGCUGCGCAGGGGGCGGGGGGCCCUGAAAAGCGGCCACU .............(((.(((..........(((((((.....)))))))((((((....))))))..))))))((((((....(((((.....))))).....))))))... ( -40.40) >DroYak_CAF1 17344 111 - 1 AAAUAAGGAUGAGUCGUCUCAGGUAGAAAGUUGCCAUUGUUUAUGGUAAUCGGCUAAAGGGGCUAAAGAGUGAGGCUGCGCAGGGGG-GUGGGACUCUGAAAAGCGGCCACU ...............(((((...(((...((((((((.....))))))))...)))..)))))..........((((((.(((((..-......)))))....))))))... ( -35.50) >DroMoj_CAF1 111706 84 - 1 UGAAAUUGGUUAGCCG-CCCAGGUAGCAAACGGCCUUUGUUUAUGGUCACAUG----------------GUGUGCCUUCUGAAGGAG-U-G-------GCGGG--AGCCACU ........(((.((..-(....)..)).)))..(((((......((.(((...----------------..)))))....)))))((-(-(-------((...--.)))))) ( -26.20) >consensus AAAUAAGGAUGAGUCGUCUCAGGUAGAAAGUUGCCAUUGUUUAUGGUAAUCGGCUAAAGG_ACUAAAGAGUGAGGCUGCGCAGGGAG_AUGGGACUCUGAAAAGCGGCCACU .........((((....))))..(((...((((((((.....))))))))...))).................((((((.(((((.........)))))....))))))... (-19.58 = -21.78 + 2.20)

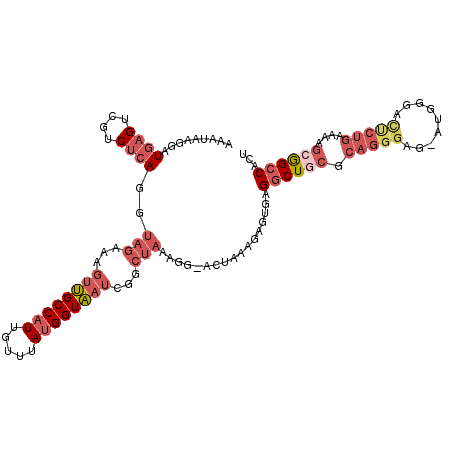
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:49 2006