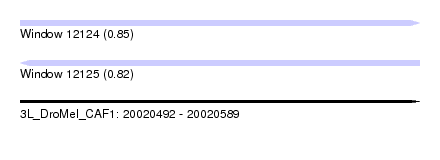
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,020,492 – 20,020,589 |
| Length | 97 |
| Max. P | 0.849453 |

| Location | 20,020,492 – 20,020,589 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -9.85 |
| Energy contribution | -9.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20020492 97 + 23771897 UAUGCAUUAUUUAUGCAAAAGUUGGUGGCCAAUAAAAUGGCAAACGGUUUGAAAAAGC-AUA-CAAGCCAAGUGCUUGGAAUGAAAAUAUCG-GUUUUGG ..(((((.....)))))((((..((((((((......)))).....((((....))))-...-(((((.....))))).........)))).-.)))).. ( -22.40) >DroVir_CAF1 12297 90 + 1 UAUGCAUAAUUUAUGCAAAACUAACAAAGCAAUAAAAUGCU-----GCAAU-AAAAGCGAUAGCAAAAUAAGUAAUGAAAAGCGAGAGAUAC----CUAC ..((((((...))))))..(((.....((((......))))-----.....-....((....))......)))...................----.... ( -10.00) >DroSec_CAF1 12261 97 + 1 UAUGCAUUAUUUAUGCAAAACUUGGUGGCCAAUAAAAUGGCAAACGGUAUCAAAAAGC-AUA-CAAACCAAGUGCUUGGAAAGGAAAUAUGG-CUUUCGG ..(((((.....)))))..(((((((.((((......)))).....((((........-)))-)..))))))).....(((((.........-))))).. ( -22.90) >DroSim_CAF1 6921 97 + 1 UAUGCAUUAUUUAUGCAAAACUUUGUGGCCAAUAAAAUGGCAAGCGGUAUGAAAAAGC-AUA-CAAACCAAGUGCUUGCAAAGGAAAUAUGG-CUUUCGG ..(((((.....)))))...((((((.((((......))))((((((((((......)-)))-)........)))))))))))((((.....-.)))).. ( -22.50) >DroEre_CAF1 11782 85 + 1 UGUGCAUUAUUUAUGCAAAACUCGGUGGCCAACAAAAUGGCAAACGCUAUGAAAG---------------AAUGCUUGUAGAGAAAAUACAGCGUUUUGG ..(((((.....)))))......((((((((......))))...)))).....((---------------((((((.(((.......))))))))))).. ( -20.00) >DroYak_CAF1 13786 98 + 1 UGUGCAUUAUUUAUGCAAAACUUGGUGGCCAAUAAAAUGGCAACCGGCAUGGAAAAGC-AUA-CAAAACAAGUGCUUGUAAAGAAAAUAUAGGGUUUUGG (((((.((.(((((((......((((.((((......)))).))))))))))).))))-)))-((((((.....((((((.......)))))))))))). ( -25.00) >consensus UAUGCAUUAUUUAUGCAAAACUUGGUGGCCAAUAAAAUGGCAAACGGUAUGAAAAAGC_AUA_CAAACCAAGUGCUUGGAAAGAAAAUAUAG_GUUUUGG ..(((((.....)))))..........((((......))))........................................................... ( -9.85 = -9.30 + -0.55)

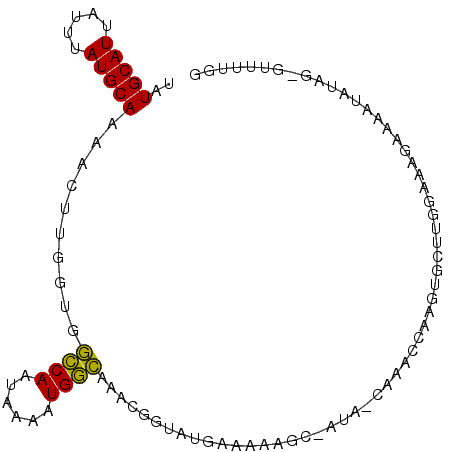
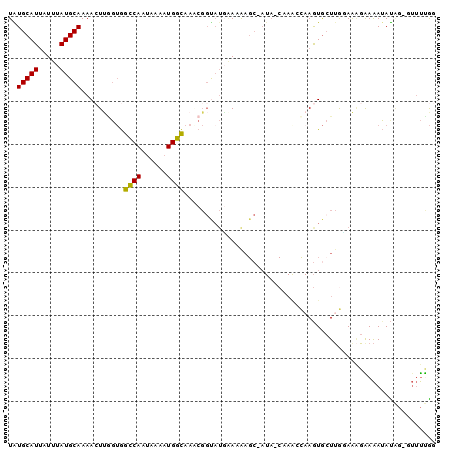
| Location | 20,020,492 – 20,020,589 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -17.28 |
| Consensus MFE | -9.32 |
| Energy contribution | -8.77 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
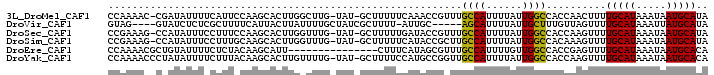
>3L_DroMel_CAF1 20020492 97 - 23771897 CCAAAAC-CGAUAUUUUCAUUCCAAGCACUUGGCUUG-UAU-GCUUUUUCAAACCGUUUGCCAUUUUAUUGGCCACCAACUUUUGCAUAAAUAAUGCAUA .......-.((......(((..(((((.....)))))-.))-).....)).........((((......))))..........(((((.....))))).. ( -15.50) >DroVir_CAF1 12297 90 - 1 GUAG----GUAUCUCUCGCUUUUCAUUACUUAUUUUGCUAUCGCUUUU-AUUGC-----AGCAUUUUAUUGCUUUGUUAGUUUUGCAUAAAUUAUGCAUA ((((----(((...............)))))))...((((.(((....-...))-----((((......))))..).))))..((((((...)))))).. ( -13.26) >DroSec_CAF1 12261 97 - 1 CCGAAAG-CCAUAUUUCCUUUCCAAGCACUUGGUUUG-UAU-GCUUUUUGAUACCGUUUGCCAUUUUAUUGGCCACCAAGUUUUGCAUAAAUAAUGCAUA ..(((((-.........))))).....(((((((..(-(((-........)))).....((((......)))).)))))))..(((((.....))))).. ( -19.40) >DroSim_CAF1 6921 97 - 1 CCGAAAG-CCAUAUUUCCUUUGCAAGCACUUGGUUUG-UAU-GCUUUUUCAUACCGCUUGCCAUUUUAUUGGCCACAAAGUUUUGCAUAAAUAAUGCAUA ......(-(..(((((....((((((.(((((((..(-(((-(......))))).))).((((......))))....)))))))))).)))))..))... ( -19.00) >DroEre_CAF1 11782 85 - 1 CCAAAACGCUGUAUUUUCUCUACAAGCAUU---------------CUUUCAUAGCGUUUGCCAUUUUGUUGGCCACCGAGUUUUGCAUAAAUAAUGCACA ...(((((((((..................---------------.....)))))))))((((......))))..........(((((.....))))).. ( -16.80) >DroYak_CAF1 13786 98 - 1 CCAAAACCCUAUAUUUUCUUUACAAGCACUUGUUUUG-UAU-GCUUUUCCAUGCCGGUUGCCAUUUUAUUGGCCACCAAGUUUUGCAUAAAUAAUGCACA .........................(((.((((((((-((.-((........)).(((.((((......)))).)))......)))).)))))))))... ( -19.70) >consensus CCAAAAC_CUAUAUUUCCUUUACAAGCACUUGGUUUG_UAU_GCUUUUUCAUACCGUUUGCCAUUUUAUUGGCCACCAAGUUUUGCAUAAAUAAUGCAUA ...........................................................((((......))))..........(((((.....))))).. ( -9.32 = -8.77 + -0.55)
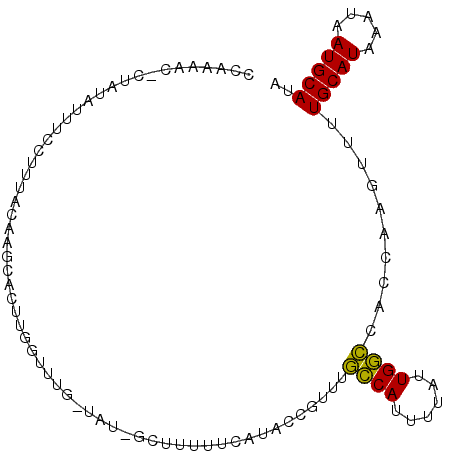
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:42 2006