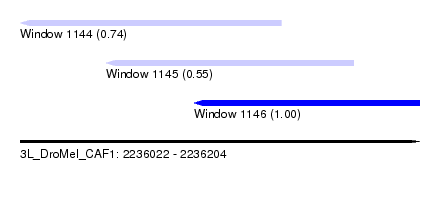
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,236,022 – 2,236,204 |
| Length | 182 |
| Max. P | 0.995130 |

| Location | 2,236,022 – 2,236,141 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -34.76 |
| Energy contribution | -33.95 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2236022 119 - 23771897 AGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAGCUUUUUGAGUCCUUCAUACUCAUGCUCACUUGGUCGUGAUCUUGGAAUGUGAGAU .(((((.(((((..(((((.((.....))((((.(.(((((.(.....).))))).).)))).)))))..))))).))))).(((((..((.(..(((....)))..)))..))))).. ( -39.40) >DroSec_CAF1 16993 119 - 1 AGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAACUUUUUGAGUCCUUUAUACUCAUGCUCACUUGGUCGUGAUCUUGGAAUGUGAGAU .(((((.(((((.((((((.....)))).((((.(.(((((.(.....).))))).).))))..))....))))).))))).(((((..((.(..(((....)))..)))..))))).. ( -34.30) >DroSim_CAF1 17027 119 - 1 AGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAGCUUUUUGAGUCCUUUAUACUCAUGCUCACUUGGUCGUGAUCUUGGAAUGUGAGAU .(((((.(((((..(((((.((.....))((((.(.(((((.(.....).))))).).)))).)))))..))))).))))).(((((..((.(..(((....)))..)))..))))).. ( -37.30) >DroEre_CAF1 30035 119 - 1 AUUGGAAGACUUUGGGGGCACGUAGCUCGGGGUGAAUGAUAGCGAAAGCGUGUCGAUGACCCAGCUCUUUGAGUCUUUUAGAUUCAUGCUCUCUUGGUCGUGUUCGUGGAAUGUGAGAU .(((((((((((..(((((.((.....))((((.(.(((((((....)).))))).).)))).)))))..))))))))))).(((((..(((..(((......))).)))..))))).. ( -43.60) >consensus AGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAGCUUUUUGAGUCCUUUAUACUCAUGCUCACUUGGUCGUGAUCUUGGAAUGUGAGAU .(((((.(((((..(((((.((.....))((((.(.(((((.(....)..))))).).)))).)))))..))))).))))).(((((..((.(..(((....)))..)))..))))).. (-34.76 = -33.95 + -0.81)

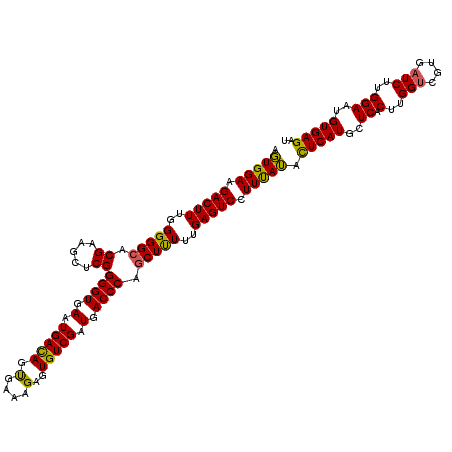
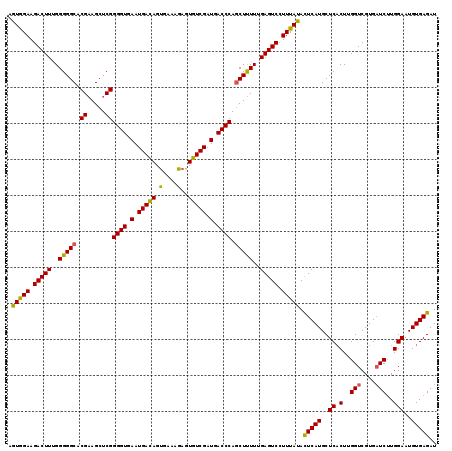
| Location | 2,236,061 – 2,236,174 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.48 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -33.67 |
| Energy contribution | -33.30 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2236061 113 - 23771897 CUCGAGCUCUCCUUCAGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAGCUUUUUGAGUCCUUCA (((((((((.((...(((.(((.((((....)))).))).).))..))))))..((((((..((((.(.(((((.(.....).))))).).)))))))))))))))....... ( -38.50) >DroSec_CAF1 17032 113 - 1 CUCGAGCUCUCCUUCUGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAACUUUUUGAGUCCUUUA ((((((((.....(((((((((...((.(((.....))).))....))))))).))))))).((((.(.(((((.(.....).))))).).))))........)))....... ( -38.20) >DroSim_CAF1 17066 113 - 1 CUCGAGCUCUCCUUCAGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAGCUUUUUGAGUCCUUUA (((((((((.((...(((.(((.((((....)))).))).).))..))))))..((((((..((((.(.(((((.(.....).))))).).)))))))))))))))....... ( -38.50) >DroEre_CAF1 30074 113 - 1 AUCGAGCUCCUCUUCAGCCUCGUCUUCUCCUGAAUUGGAAGACUUUGGGGGCACGUAGCUCGGGGUGAAUGAUAGCGAAAGCGUGUCGAUGACCCAGCUCUUUGAGUCUUUUA ..((((((.(......((((((((((((........)))))))....)))))..).))))))((((.(.(((((((....)).))))).).)))).((((...))))...... ( -39.50) >consensus CUCGAGCUCUCCUUCAGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGACAGUGAAAGAGUGUCGAUGACCCAGCUUUUUGAGUCCUUUA ..(((((((((((..(((.(((.((((....)))).))).).))..))))).....))))))((((.(.(((((.(....)..))))).).)))).((((...))))...... (-33.67 = -33.30 + -0.37)

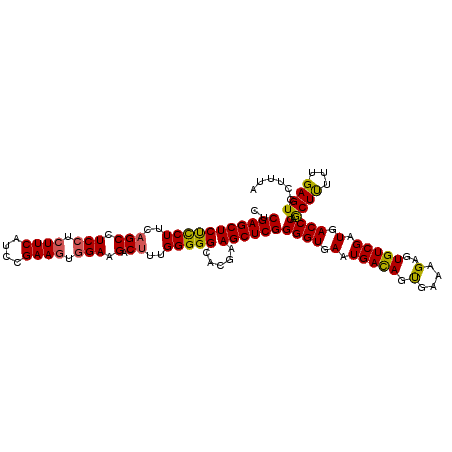
| Location | 2,236,101 – 2,236,204 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -31.08 |
| Energy contribution | -31.95 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2236101 103 - 23771897 CUCCAGUUCAUCGCCUUCAGAUACAUUUUCCUCGAGCUCUCCUUCAGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGAC .........(((.......))).(((((.((((((((((((((..(((.(((.((((....)))).))).).))..))))).....))))))))).))))).. ( -36.00) >DroSec_CAF1 17072 103 - 1 CUCCAGCUCAUUACCCUCAGAUUCAUUUUCCUCGAGCUCUCCUUCUGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGAC ......................((((((.(((((((((.....(((((((((...((.(((.....))).))....))))))).))))))))))).)))))). ( -37.40) >DroSim_CAF1 17106 103 - 1 CUCCAGCUCAUUUCCCUCAGAUUCAUUUUCCUCGAGCUCUCCUUCAGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGAC ......................((((((.((((((((((((((..(((.(((.((((....)))).))).).))..))))).....))))))))).)))))). ( -36.70) >DroEre_CAF1 30114 103 - 1 CUCCAAGUCAUCCUCUUCAUACUCAUUUACAUCGAGCUCCUCUUCAGCCUCGUCUUCUCCUGAAUUGGAAGACUUUGGGGGCACGUAGCUCGGGGUGAAUGAU ......................((((((((..((((((.(......((((((((((((........)))))))....)))))..).))))))..)))))))). ( -32.00) >consensus CUCCAGCUCAUCACCCUCAGAUUCAUUUUCCUCGAGCUCUCCUUCAGCCUCCUCUUCAUCCGAAGUGGAAGACUUUGGGGGCACGAAGCUCGGGGUGAAUGAC ......................((((((.((((((((((((((..(((.(((.((((....)))).))).).))..))))).....))))))))).)))))). (-31.08 = -31.95 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:06 2006