| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,016,828 – 20,016,933 |
| Length | 105 |
| Max. P | 0.997949 |

| Location | 20,016,828 – 20,016,933 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -22.40 |
| Energy contribution | -25.28 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20016828 105 + 23771897 ----------AUUCAAUGGAACUCUGCAUACAAAUUUUCUAUUUAGCUGCCCUUUCAAUUGAAAGGUAGAGUAUCUUCUGCGAAUCUCGGAGCCCGUUUGUGUGCAAAGUUCCCU ----------.......((((((.(((((((((((...........(((((.((((....)))))))))........((.((.....)).))...))))))))))).)))))).. ( -34.20) >DroSec_CAF1 8746 105 + 1 ----------AUUCAACGGCACUCUGCACACAAAUUUUCUAUUGGGCUGUCCUUUCAAUUGAAAGGUAGAGUAUCUCCUGCGAAUCUCGAGGCCCGUUUGUGUGCAAAGUUCCCU ----------.......((.(((.(((((((((((........(((((..((((((....))))))..(((..((......))..)))..)))))))))))))))).))).)).. ( -37.30) >DroSim_CAF1 3421 105 + 1 ----------AUUCAAUGGCACUCUGCAUACAAAUUUUCUAUUGGGCUGCCCUUUCAAUUGAAAGGUAGAGUAUUUCCUGCGAAUCUCGAGGCCCGUUUGUGUGCAAAGUUCCCU ----------.......((.(((.(((((((((((........(((((..((((((....))))))..(((...(((....))).)))..)))))))))))))))).))).)).. ( -35.60) >DroYak_CAF1 10202 106 + 1 CUCCCUUUUCAUUCAAUGACACUUUGAAUAUUUAUUUUCUAUUGGGUUGCCCAUGCAAUUGAAAGGUAGAGUAUCUCCUGCGAAUCUCGGAGCCCGUCUG--------U-UCCCU ....((((((((((((.......))))))...............((((((....))))))))))))..(((..((......))..)))(((((......)--------)-))).. ( -20.10) >consensus __________AUUCAAUGGCACUCUGCAUACAAAUUUUCUAUUGGGCUGCCCUUUCAAUUGAAAGGUAGAGUAUCUCCUGCGAAUCUCGAAGCCCGUUUGUGUGCAAAGUUCCCU .................((.(((.(((((((((((........(((((.(((((((....))))))..(((..((......))..)))).)))))))))))))))).))).)).. (-22.40 = -25.28 + 2.88)

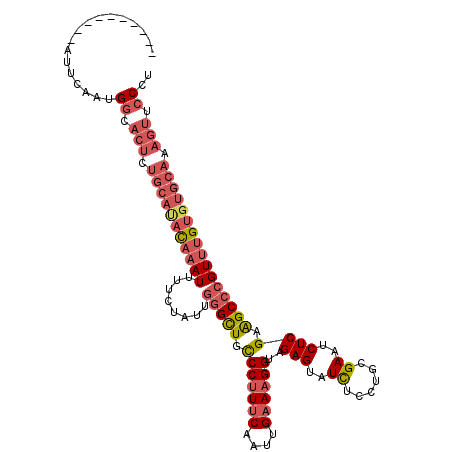
| Location | 20,016,828 – 20,016,933 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -18.40 |
| Energy contribution | -22.27 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20016828 105 - 23771897 AGGGAACUUUGCACACAAACGGGCUCCGAGAUUCGCAGAAGAUACUCUACCUUUCAAUUGAAAGGGCAGCUAAAUAGAAAAUUUGUAUGCAGAGUUCCAUUGAAU---------- ..(((((((((((.(((((..((((.((((..((......))..)))..((((((....))))))).))))..........))))).))))))))))).......---------- ( -34.90) >DroSec_CAF1 8746 105 - 1 AGGGAACUUUGCACACAAACGGGCCUCGAGAUUCGCAGGAGAUACUCUACCUUUCAAUUGAAAGGACAGCCCAAUAGAAAAUUUGUGUGCAGAGUGCCGUUGAAU---------- ..((.((((((((((((((.((((...(((..((......))..)))..((((((....))))))...)))).........)))))))))))))).)).......---------- ( -38.90) >DroSim_CAF1 3421 105 - 1 AGGGAACUUUGCACACAAACGGGCCUCGAGAUUCGCAGGAAAUACUCUACCUUUCAAUUGAAAGGGCAGCCCAAUAGAAAAUUUGUAUGCAGAGUGCCAUUGAAU---------- ..((.((((((((.(((((.((((...(((.(((....)))...)))..((((((....))))))...)))).........))))).)))))))).)).......---------- ( -31.80) >DroYak_CAF1 10202 106 - 1 AGGGA-A--------CAGACGGGCUCCGAGAUUCGCAGGAGAUACUCUACCUUUCAAUUGCAUGGGCAACCCAAUAGAAAAUAAAUAUUCAAAGUGUCAUUGAAUGAAAAGGGAG .(...-.--------)(((.(..((((..........))))...)))).(((((((((.((((((....)).....(((........)))...)))).))))).....))))... ( -20.00) >consensus AGGGAACUUUGCACACAAACGGGCCCCGAGAUUCGCAGGAGAUACUCUACCUUUCAAUUGAAAGGGCAGCCCAAUAGAAAAUUUGUAUGCAGAGUGCCAUUGAAU__________ ..((.((((((((.(((((.((((...(((..((......))..)))..((((((....))))))...)))).........))))).)))))))).))................. (-18.40 = -22.27 + 3.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:38 2006