| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,014,883 – 20,014,975 |
| Length | 92 |
| Max. P | 0.859688 |

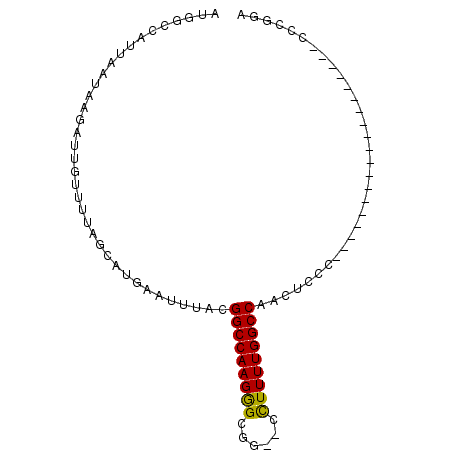
| Location | 20,014,883 – 20,014,975 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20014883 92 + 23771897 AUGGCCAUUAAUAAGAUUGUUUUAGCAAGAAUUUACGGCCAAGGGCGGGCCUUUUUGGCCAACUCCUCGCCCCUGGUUCGUUGAUUCCCUGG ....(((.........((((....))))(((((.(((((((.(((((((((.....)))).......))))).))).)))).)))))..))) ( -30.61) >DroSec_CAF1 6893 71 + 1 AUGGCCAUUAAUAAGAUUGUUUUAUCAUGAAUUUACGGCCAAGGGCGG--CCUUUUGGCCAACUCCC-------------------CCCGUA .(((((........(((......)))..(......)))))).(((.((--((....))))....)))-------------------...... ( -19.00) >DroSim_CAF1 695 71 + 1 AUGGCCAUUAAUAAGAUUGUUUUAGCAUGAAUUUACGGCCAAGAGCGG--CCUUUUGGCCAACUCCC-------------------CCCGGA .((((((..........(((....))).........((((......))--))...))))))..(((.-------------------...))) ( -18.60) >DroYak_CAF1 8260 70 + 1 AUGGCCAUUAAUAAGAUUGUUUUAGCAUGAAUUUACGGCCAAGGGCGGGCCUUUUUGGCC---UCUC-------------------GUCUAU .(((((..(((......(((....))).....))).))))).(((((((((.....))))---...)-------------------)))).. ( -20.60) >consensus AUGGCCAUUAAUAAGAUUGUUUUAGCAUGAAUUUACGGCCAAGGGCGG__CCUUUUGGCCAACUCCC___________________CCCGGA ....................................(((((((((......)))))))))................................ (-12.66 = -12.22 + -0.44)


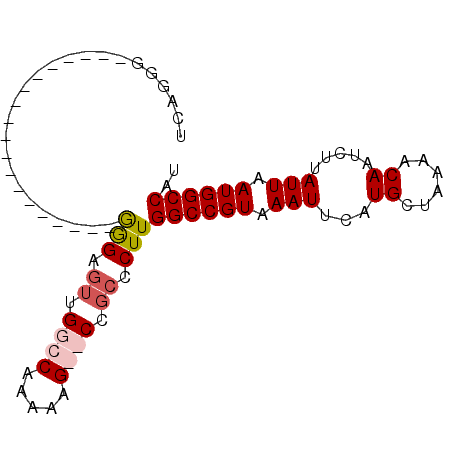
| Location | 20,014,883 – 20,014,975 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -23.39 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.82 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20014883 92 - 23771897 CCAGGGAAUCAACGAACCAGGGGCGAGGAGUUGGCCAAAAAGGCCCGCCCUUGGCCGUAAAUUCUUGCUAAAACAAUCUUAUUAAUGGCCAU ...((((((..(((..(((((((((.......((((.....))))))))))))).)))..))))))((((...............))))... ( -29.97) >DroSec_CAF1 6893 71 - 1 UACGGG-------------------GGGAGUUGGCCAAAAGG--CCGCCCUUGGCCGUAAAUUCAUGAUAAAACAAUCUUAUUAAUGGCCAU ....((-------------------(((....((((....))--)).)))))((((((.(((....(((......)))..))).)))))).. ( -24.60) >DroSim_CAF1 695 71 - 1 UCCGGG-------------------GGGAGUUGGCCAAAAGG--CCGCUCUUGGCCGUAAAUUCAUGCUAAAACAAUCUUAUUAAUGGCCAU ......-------------------((((((.((((....))--))))))))((((((.(((...((......)).....))).)))))).. ( -22.30) >DroYak_CAF1 8260 70 - 1 AUAGAC-------------------GAGA---GGCCAAAAAGGCCCGCCCUUGGCCGUAAAUUCAUGCUAAAACAAUCUUAUUAAUGGCCAU ......-------------------....---(((((....((((.......))))(((......))).................))))).. ( -16.70) >consensus UCAGGG___________________GGGAGUUGGCCAAAAAG__CCGCCCUUGGCCGUAAAUUCAUGCUAAAACAAUCUUAUUAAUGGCCAU .........................(((.((.((((.....)))).)).)))((((((.(((...((......)).....))).)))))).. (-11.70 = -12.82 + 1.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:34 2006