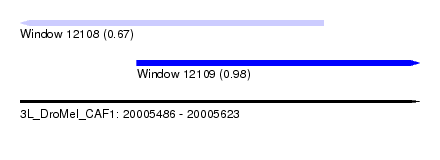
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,005,486 – 20,005,623 |
| Length | 137 |
| Max. P | 0.976292 |

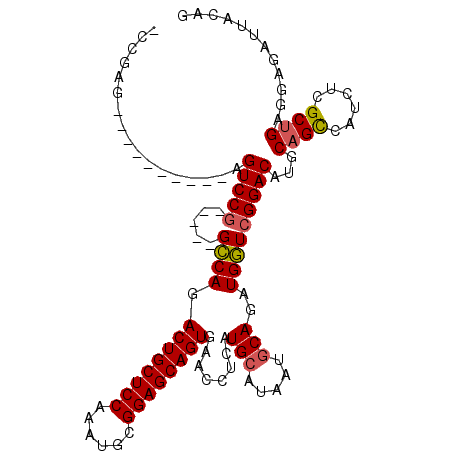
| Location | 20,005,486 – 20,005,590 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20005486 104 - 23771897 GACCAUCUGCAUUAUGCAUGAGGUUCACUGCUCCGCAUUUGGAGCAGUCUGGC-----AGGACU----------CUCGG-CAUCCUUUUGUAAGCAAGUGCGAAAGUUUUAUUUAACAAU (((..((.(((((.(((.(((((((((((((((((....))))))))).....-----.)))).----------))))(-((......)))..))))))))))..)))............ ( -30.20) >DroSec_CAF1 95737 104 - 1 GACCAUCUGCAUCAUCCAUGAGGUUCACUGCUCCGUAUUUGGAGCAGUCUGGC-----CGGACU----------CACGG-CAUCCUUUUGUUGGCAAGUGCGAAACUUUUAUUUAACUAU .......(((((...(((.((((...(((((((((....)))))))))...((-----((....----------..)))-)..))))....)))...))))).................. ( -33.10) >DroSim_CAF1 54713 104 - 1 GACCAUCUGCAUUAUGCAUGAGGUUCACUGCUCCGCAUUUGGAGCAGUCUGGC-----CGGACU----------CACGG-CAUCCUUUUGUUGGCAAGUGCGAAACUUUUAUUUAACUAU ..(((..(((.....(((.((((...(((((((((....)))))))))...((-----((....----------..)))-)..)))).)))..)))..)).).................. ( -32.90) >DroEre_CAF1 46062 120 - 1 GAGCAACUGCAUUAUGCAUGAGGUUCACUGCUCCGCAUUUGGAGCAGUCUGCCGCACUCGGACUCGGACUCGAACUCGGACAGCCUUUUGUGAGCAAGUGGGAAGCUUUUAUUUAACAAU ((((..((.((((.(((....(((..(((((((((....)))))))))..))).(((..((..((.((.......)).))...))....))).)))))))))..))))............ ( -34.30) >DroYak_CAF1 51928 104 - 1 GACCAACUGCAUUAUGCAUGAGGUUCACUGCUCCGCAUUUGGAGCAGUCUGGC-----CGGACU----------CUCGG-CAGCCUUUUGUAAGCAAGUGAGAAACUUGUAUUUAACAAU ..............((((.((((((.(((((((((....)))))))))...((-----(((...----------.))))-))))))).)))).((((((.....)))))).......... ( -36.40) >consensus GACCAUCUGCAUUAUGCAUGAGGUUCACUGCUCCGCAUUUGGAGCAGUCUGGC_____CGGACU__________CUCGG_CAUCCUUUUGUAAGCAAGUGCGAAACUUUUAUUUAACAAU ...((..(((....((((.((((.....(((((((....)))))))(((((.......)))))....................)))).)))).)))..)).................... (-23.30 = -24.02 + 0.72)

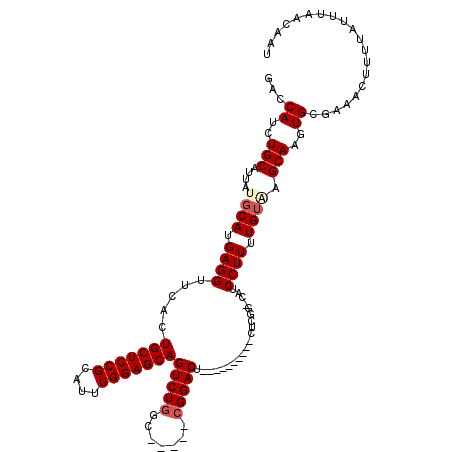
| Location | 20,005,526 – 20,005,623 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -29.48 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20005526 97 + 23771897 -CCGAG----------AGUCCU-----GCCAGACUGCUCCAAAUGCGGAGCAGUGAACCUCAUGCAUAAUGCAGAUGGUCGGACAUGCAGCCAUCUCGCUGAGGAGAUUACAG -((...----------.((((.-----((((.((((((((......)))))))).....((.(((.....))))))))).))))...((((......)))).))......... ( -35.60) >DroSec_CAF1 95777 97 + 1 -CCGUG----------AGUCCG-----GCCAGACUGCUCCAAAUACGGAGCAGUGAACCUCAUGGAUGAUGCAGAUGGUCGGACAUGCAGUCAUCUCGCUGAGCAGAUUACAG -..(((----------.(((((-----((((.((((((((......))))))))...((....))..........))))))))).))).((.((((.((...)))))).)).. ( -38.40) >DroSim_CAF1 54753 97 + 1 -CCGUG----------AGUCCG-----GCCAGACUGCUCCAAAUGCGGAGCAGUGAACCUCAUGCAUAAUGCAGAUGGUCGGACAUGCAGCCAUCUCGCUGAGCAGAUUACAG -..(((----------((((((-----((((.((((((((......)))))))).....((.(((.....))))))))))))))...((((......))))......)))).. ( -41.40) >DroEre_CAF1 46102 113 + 1 UCCGAGUUCGAGUCCGAGUCCGAGUGCGGCAGACUGCUCCAAAUGCGGAGCAGUGAACCUCAUGCAUAAUGCAGUUGCUCGGACUUGCAGCCAUCUCGCCGAGGAGAUUACAG ...(.((((((((((((((....(((.((...((((((((......))))))))...)).)))((.....))....))))))))))).))))(((((......)))))..... ( -44.20) >DroYak_CAF1 51968 97 + 1 -CCGAG----------AGUCCG-----GCCAGACUGCUCCAAAUGCGGAGCAGUGAACCUCAUGCAUAAUGCAGUUGGUCGGACAUGCAGCCAACUCGCUGAAGAGAUUACCG -.....----------.(((((-----(((((((((((((......))))))))........(((.....))).))))))))))...((((......))))............ ( -39.10) >consensus _CCGAG__________AGUCCG_____GCCAGACUGCUCCAAAUGCGGAGCAGUGAACCUCAUGCAUAAUGCAGAUGGUCGGACAUGCAGCCAUCUCGCUGAGGAGAUUACAG .................(((((.....((((.((((((((......))))))))........(((.....)))..)))))))))...((((......))))............ (-29.48 = -29.60 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:27 2006