
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,004,661 – 20,004,797 |
| Length | 136 |
| Max. P | 0.996151 |

| Location | 20,004,661 – 20,004,759 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20004661 98 + 23771897 UUGAUUGGGGGGUCUGGAAAAGUGGCAAAUGCAUUUGGGCCAUUCGACCAUUCGGCCCAAAUGCGAAUUUACGAAUUCAAAUUCGAUUGGGUAAUAAA (..((((((..(((.........)))....(((((((((((............)))))))))))(((((....)))))...))))))..)........ ( -28.40) >DroSec_CAF1 94939 89 + 1 UUGAUUGGG-GCUGUGGAAAAGUGGCAAAUGCAUUUGGGCCAUUC--------GACCCAAAUGCGGAUUUUCGAAUUCAAAUUCGAUUGGGUAAUAAA (..((((((-((..(......)..))....(((((((((......--------..)))))))))(((((....)))))...))))))..)........ ( -24.20) >DroSim_CAF1 53859 89 + 1 UUGAUUGGG-GUUGUGGAAAAGUGGCAAAUGCAUUUGGGCCAUUC--------GGCCCAAAUGCGGAUUUUCGAAUUCAAAUUCGAUUGGGUAAUAAA (..((((((-((..(......)..))....(((((((((((....--------)))))))))))(((((....)))))...))))))..)........ ( -29.80) >DroYak_CAF1 51078 85 + 1 UUGAUUGGG-GGGCUUGGAAAAUGGGAAAUGCAUU---GCCAUUC--------GGCCCAAAUGCGGAU-UGCGAAUUCAAAUUCGAUCGGGGACUAAA ..(((((((-(((((.....(((((.((.....))---.))))).--------)))))...(((....-.)))........)))))))(....).... ( -20.50) >consensus UUGAUUGGG_GGUCUGGAAAAGUGGCAAAUGCAUUUGGGCCAUUC________GGCCCAAAUGCGGAUUUUCGAAUUCAAAUUCGAUUGGGUAAUAAA (((((((((.....................(((((((((((............)))))))))))(((((....)))))...)))))))))........ (-20.08 = -20.95 + 0.87)

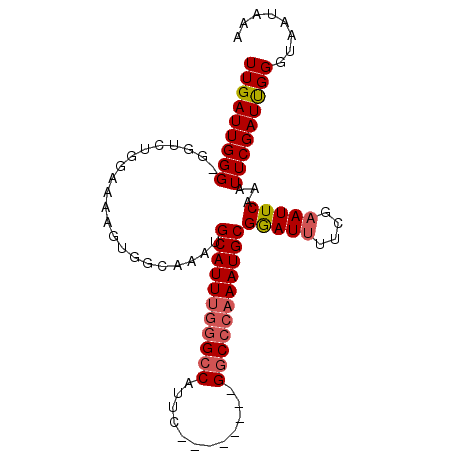

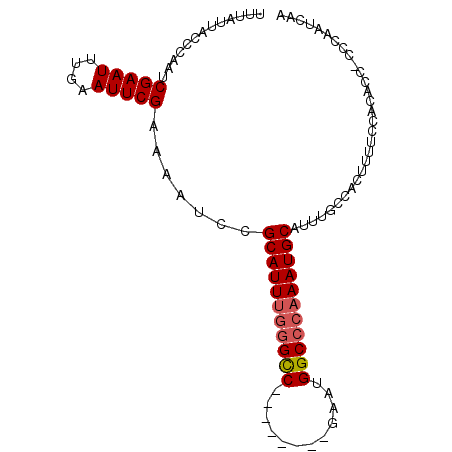
| Location | 20,004,661 – 20,004,759 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20004661 98 - 23771897 UUUAUUACCCAAUCGAAUUUGAAUUCGUAAAUUCGCAUUUGGGCCGAAUGGUCGAAUGGCCCAAAUGCAUUUGCCACUUUUCCAGACCCCCCAAUCAA ..............(((((((......)))))))((((((((((((..........)))))))))))).............................. ( -24.60) >DroSec_CAF1 94939 89 - 1 UUUAUUACCCAAUCGAAUUUGAAUUCGAAAAUCCGCAUUUGGGUC--------GAAUGGCCCAAAUGCAUUUGCCACUUUUCCACAGC-CCCAAUCAA ............((((((....))))))......(((((((((((--------....)))))))))))....................-......... ( -23.10) >DroSim_CAF1 53859 89 - 1 UUUAUUACCCAAUCGAAUUUGAAUUCGAAAAUCCGCAUUUGGGCC--------GAAUGGCCCAAAUGCAUUUGCCACUUUUCCACAAC-CCCAAUCAA ............((((((....))))))......(((((((((((--------....)))))))))))....................-......... ( -25.80) >DroYak_CAF1 51078 85 - 1 UUUAGUCCCCGAUCGAAUUUGAAUUCGCA-AUCCGCAUUUGGGCC--------GAAUGGC---AAUGCAUUUCCCAUUUUCCAAGCCC-CCCAAUCAA ..........((((((((....)))))..-))).......((((.--------((((((.---((.....)).)))))).....))))-......... ( -12.90) >consensus UUUAUUACCCAAUCGAAUUUGAAUUCGAAAAUCCGCAUUUGGGCC________GAAUGGCCCAAAUGCAUUUGCCACUUUUCCACACC_CCCAAUCAA .............(((((....))))).......(((((((((((............))))))))))).............................. (-18.14 = -18.70 + 0.56)

| Location | 20,004,684 – 20,004,797 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -12.66 |
| Energy contribution | -15.26 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20004684 113 + 23771897 -----GGCAAAUGCAUUUGGGCCAUUCGACCAUUCGGCCCAAAUGCGAAUUUACGAAUUCAAAUUCGAUUGGGUAAUAAAACAAUGU--GCAUAAAAGGUUAACGAUUGGCAAUAAAAUU -----.((....(((((((((((............)))))))))))(((((....))))).....((((((..((((..........--.........)))).))))))))......... ( -28.01) >DroSec_CAF1 94961 98 + 1 -----GGCAAAUGCAUUUGGGCCAUUC--------GACCCAAAUGCGGAUUUUCGAAUUCAAAUUCGAUUGGGUAAUAAAACAAUGU--GCAUAAAAGGUUAACGAUUGGAAA------- -----.(((..((((((((((......--------..)))))))))).....((((((....))))))..................)--))......................------- ( -21.90) >DroSim_CAF1 53881 105 + 1 -----GGCAAAUGCAUUUGGGCCAUUC--------GGCCCAAAUGCGGAUUUUCGAAUUCAAAUUCGAUUGGGUAAUAAAACAAUGU--GCAUAAAAGGUUAACGAUUGGCAAUAAAAUC -----.(((..((((((((((((....--------)))))))))))).....((((((....))))))..................)--)).............((((........)))) ( -29.50) >DroYak_CAF1 51100 101 + 1 -----GGGAAAUGCAUU---GCCAUUC--------GGCCCAAAUGCGGAU-UGCGAAUUCAAAUUCGAUCGGGGACUAAAACAAAGU--ACAUAAAAGGUUUACGGGUGGCAAUAAAAUC -----.........(((---(((((((--------(.(((.....(((((-(..(....).))))))....)))(((.......)))--..............)))))))))))...... ( -25.40) >DroAna_CAF1 49326 103 + 1 UCCACGCAAAAUGCA--------AUUC--------ACUCUGAACGCGUAU-UUCGAAUUCAAAUUCGUUGCGAAAAUAAAACAGAGUUCACAUAAAAAGUUAACGAGAAACAGUCAAAAC .....((.....)).--------....--------((((((....((((.-..(((((....))))).)))).........))))))................................. ( -13.62) >consensus _____GGCAAAUGCAUUUGGGCCAUUC________GGCCCAAAUGCGGAUUUUCGAAUUCAAAUUCGAUUGGGUAAUAAAACAAUGU__GCAUAAAAGGUUAACGAUUGGCAAUAAAAUC ...........((((((((((((............))))))))))))......(((((....)))))..................................................... (-12.66 = -15.26 + 2.60)

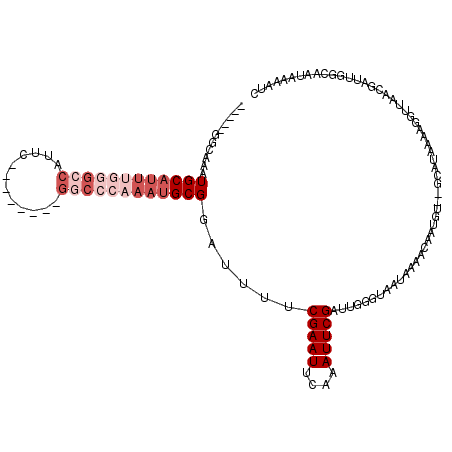
| Location | 20,004,684 – 20,004,797 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -13.68 |
| Energy contribution | -15.60 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
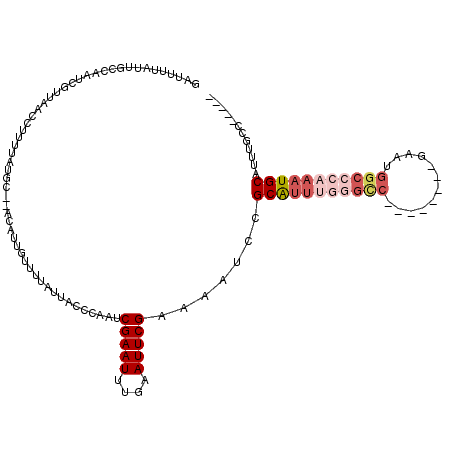
>3L_DroMel_CAF1 20004684 113 - 23771897 AAUUUUAUUGCCAAUCGUUAACCUUUUAUGC--ACAUUGUUUUAUUACCCAAUCGAAUUUGAAUUCGUAAAUUCGCAUUUGGGCCGAAUGGUCGAAUGGCCCAAAUGCAUUUGCC----- .............................((--(....................(((((((......)))))))((((((((((((..........))))))))))))...))).----- ( -26.50) >DroSec_CAF1 94961 98 - 1 -------UUUCCAAUCGUUAACCUUUUAUGC--ACAUUGUUUUAUUACCCAAUCGAAUUUGAAUUCGAAAAUCCGCAUUUGGGUC--------GAAUGGCCCAAAUGCAUUUGCC----- -------......................((--(..................((((((....))))))......(((((((((((--------....)))))))))))...))).----- ( -25.00) >DroSim_CAF1 53881 105 - 1 GAUUUUAUUGCCAAUCGUUAACCUUUUAUGC--ACAUUGUUUUAUUACCCAAUCGAAUUUGAAUUCGAAAAUCCGCAUUUGGGCC--------GAAUGGCCCAAAUGCAUUUGCC----- ((((........)))).............((--(..................((((((....))))))......(((((((((((--------....)))))))))))...))).----- ( -28.30) >DroYak_CAF1 51100 101 - 1 GAUUUUAUUGCCACCCGUAAACCUUUUAUGU--ACUUUGUUUUAGUCCCCGAUCGAAUUUGAAUUCGCA-AUCCGCAUUUGGGCC--------GAAUGGC---AAUGCAUUUCCC----- .....((((((((..(((((.....))))).--..............(((((..((((....))))((.-....))..)))))..--------...))))---))))........----- ( -18.50) >DroAna_CAF1 49326 103 - 1 GUUUUGACUGUUUCUCGUUAACUUUUUAUGUGAACUCUGUUUUAUUUUCGCAACGAAUUUGAAUUCGAA-AUACGCGUUCAGAGU--------GAAU--------UGCAUUUUGCGUGGA (((((((.........(((.((.......)).))).....((((..((((...))))..)))).)))))-)).((((..((((((--------(...--------..))))))))))).. ( -17.30) >consensus GAUUUUAUUGCCAAUCGUUAACCUUUUAUGC__ACAUUGUUUUAUUACCCAAUCGAAUUUGAAUUCGAAAAUCCGCAUUUGGGCC________GAAUGGCCCAAAUGCAUUUGCC_____ .....................................................(((((....))))).......(((((((((((............)))))))))))............ (-13.68 = -15.60 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:25 2006