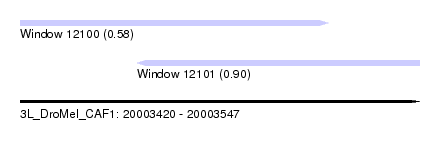
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,003,420 – 20,003,547 |
| Length | 127 |
| Max. P | 0.899779 |

| Location | 20,003,420 – 20,003,518 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.84 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -13.23 |
| Energy contribution | -12.95 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20003420 98 + 23771897 GGCACCAGCACUCUACCAGGCAAACACUGGCAACACA---------UACAUAUUGGGCUAGCUUGCUGGCCAAACACUUUGAAUGGAGUGCG-------------AAUUGCAUUGGUUGA .(((...((((((((((((.......)))).......---------.........((((((....))))))............)))))))).-------------...)))......... ( -31.20) >DroSec_CAF1 93787 79 + 1 GGCAUCAGCAAUCGACC-------------------A---------UACAUAUAUGGCUGGCUUUCUGGCCAAACACUUUGAAUUAAGUGCG-------------AGUUGCAUUGGUUGA .((....))..((((((-------------------(---------........((((..(....)..))))..(((((......)))))..-------------........))))))) ( -21.30) >DroSim_CAF1 52714 79 + 1 GGCAACAGCACUCUACC-------------------A---------UACAUAUAUGGCUGGCUUUCUGGCCAAACACUUUGAAUUAAGUGCG-------------AAUUGCAUUGCUUGA .((((..(((((...((-------------------(---------((....))))).(((((....)))))..............))))).-------------..))))......... ( -21.00) >DroEre_CAF1 44141 116 + 1 GGCAUCGGCACUCUGGCAGCCAAACACUGGCCACACAUACGA---GUGUGUAUAUCACUGGCUUUCCAGCCAAACACUUUGAAUUGAGUGCC-AGGCAAGGAUGUAGCUGCAUUGGUAGA .((((((((((((...((((((.....))))(((((......---)))))........(((((....))))).......))....)))))))-.......)))))..((((....)))). ( -35.11) >DroYak_CAF1 49728 120 + 1 GGCAUCAGCACUCUGGCAGCCAAAAACUAUAAACACAUAUUAUACAUAUAUGUAUGGCUGGCUUUCCAACCAAACACUUUGAAUUGAGUGCUAAGGCAAGGUCGUAGCUGCAUUGGUGGA ..((((((((((.((((.(((.............((((((.......)))))).(((.(((....))).)))..(((((......)))))....)))...)))).)).)))..))))).. ( -29.40) >DroAna_CAF1 48178 108 + 1 GGCUCCGGCACUUUAGCGGGAAAA---UAGACACACACAUUA---------AUACGGCUGGCUUUCUGUCCAAACACUCUGUAUUGAGUGAGAAAGUGAGGACGUAAUUGCAUUGGCUAG ((((((.((......)).)))...---...............---------.((((.((.(((((((.......(((((......)))))))))))).))..)))).........))).. ( -28.71) >consensus GGCAUCAGCACUCUACCAG_CAAA___U_G__ACACA_________UACAUAUAUGGCUGGCUUUCUGGCCAAACACUUUGAAUUGAGUGCG_____________AAUUGCAUUGGUUGA .......(((.............................................((((((....))))))...(((((......)))))..................)))......... (-13.23 = -12.95 + -0.28)



| Location | 20,003,457 – 20,003,547 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.45 |
| Mean single sequence MFE | -15.87 |
| Consensus MFE | -9.68 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20003457 90 - 23771897 AAUUCCCUUGACUUAACAU--C------GAAACCCAUUCAACCAAUGCAAUU------------CGCACUCCAUUCAAAGUGUUUGGCCAGCAAGCUAGCCCAAUAUGUA--- .......((((........--(------(((...((((.....))))...))------------))........)))).(((((.(((.((....)).))).)))))...--- ( -12.29) >DroSec_CAF1 93805 77 - 1 AAUUCCCU---------AU--C----------CAUAUUCAACCAAUGCAACU------------CGCACUUAAUUCAAAGUGUUUGGCCAGAAAGCCAGCCAUAUAUGUA--- ........---------..--.----------(((((...............------------.((((((......))))))(((((......)))))....)))))..--- ( -11.70) >DroSim_CAF1 52732 77 - 1 AAUUCCCU---------AU--C----------CAUAUUCAAGCAAUGCAAUU------------CGCACUUAAUUCAAAGUGUUUGGCCAGAAAGCCAGCCAUAUAUGUA--- ........---------..--.----------(((((....((.........------------.((((((......)))))).((((......))))))...)))))..--- ( -14.20) >DroEre_CAF1 44181 113 - 1 AAUUUCCUUAAUUCACCAUGCCCACGAGGAAGCCAAUUCUACCAAUGCAGCUACAUCCUUGCCUGGCACUCAAUUCAAAGUGUUUGGCUGGAAAGCCAGUGAUAUACACACUC ............((((..((((..(((((((((..(((.....)))...)))...))))))...))))................(((((....)))))))))........... ( -25.30) >consensus AAUUCCCU_________AU__C__________CAUAUUCAACCAAUGCAACU____________CGCACUCAAUUCAAAGUGUUUGGCCAGAAAGCCAGCCAUAUAUGUA___ ..............................................((.................((((((......)))))).((((......))))))............. ( -9.68 = -9.80 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:19 2006