
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,003,252 – 20,003,344 |
| Length | 92 |
| Max. P | 0.852707 |

| Location | 20,003,252 – 20,003,344 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
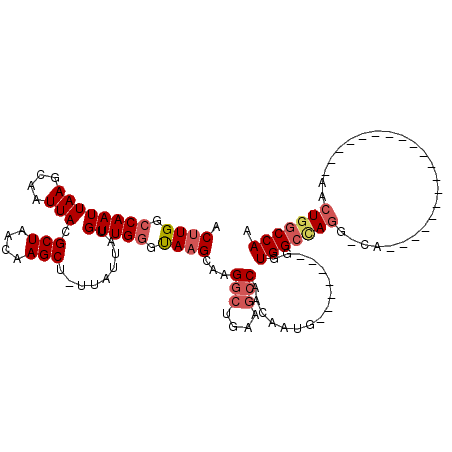
>3L_DroMel_CAF1 20003252 92 + 23771897 UUGGCCAGUU-------------------UG-CCUAGCCACC-------CAUUGUUGGCUUCAGCCUUGCUUACCCAACAUAAUAA-AGCUUGUUAGCGUAAUUGCUUAAUUGGCCAAGU ((((((((((-------------------..-...(((....-------...((((((....(((...)))...))))))......-.(((....)))......))).)))))))))).. ( -25.90) >DroSec_CAF1 93612 99 + 1 UUGGCCAGUU-------------------UG-CCUGGCCACCCAUUACCCAUUGUUGGCUUCAGCCUUGCUUACCCAACAUAAUAA-AGCUUGUUAGCGUAAUUGCUUAAUUGGCCAAGU ((((((((((-------------------.(-(..((....))(((((....((((((....(((...)))...))))))......-.(((....)))))))).))..)))))))))).. ( -27.80) >DroSim_CAF1 52539 99 + 1 UUGGCCAGUU-------------------UG-CCUGGCCACCCAUUACCCAUUGUUGGCUUCAGCCUUGCUUACCCAACAUAAUAA-AGCUUGUUAGCGUAAUUGCUUAAUUGGCCAAGU ((((((((((-------------------.(-(..((....))(((((....((((((....(((...)))...))))))......-.(((....)))))))).))..)))))))))).. ( -27.80) >DroEre_CAF1 43974 91 + 1 UUGGCCAGU--------------------UG-ACUGCCCACC-------CAUUGUUGGCUUCAGCCUUGCUUGCCCAACAUAAUAA-AGCUUGUUAGCGUAAUUGCUUAAUUGGCCAAGU (((((((((--------------------((-(..((.....-------...(((((((...(((...))).).))))))......-.(((....)))......)))))))))))))).. ( -27.10) >DroYak_CAF1 49562 90 + 1 UUGGCCAGU--------------------UG-ACUAGCCACC-------CAUUGUUGGC-UCAGUCUUGCUUACCCAACAUAAUAA-AGCUUGUUAGCGUAAUUGCUUAAUUGGCCAAGU (((((((((--------------------((-((((((((..-------......))))-).))))..(((((((.((((......-....)))).).))))..))..)))))))))).. ( -28.90) >DroAna_CAF1 47983 115 + 1 UUGGCUAGUUCCUGGGUGGGUCACGGGAUUGACUUGGCCAU-----GGCCAUUGUUGGUUUCAGCCCGGCUUGCACAACAUAAUAAAAGCUUGUUAGCGUAAUUGCUUAAUUGGCCAGGA ((((((((((...((.(((((...(..((..((.((((...-----.))))..))..))..).))))).)).(((..((.((((((....))))))..))...)))..)))))))))).. ( -39.20) >consensus UUGGCCAGUU___________________UG_CCUGGCCACC_______CAUUGUUGGCUUCAGCCUUGCUUACCCAACAUAAUAA_AGCUUGUUAGCGUAAUUGCUUAAUUGGCCAAGU ((((((((((..........................................((((((....(((...)))...))))))........(((....)))..........)))))))))).. (-21.50 = -21.58 + 0.09)


| Location | 20,003,252 – 20,003,344 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20003252 92 - 23771897 ACUUGGCCAAUUAAGCAAUUACGCUAACAAGCU-UUAUUAUGUUGGGUAAGCAAGGCUGAAGCCAACAAUG-------GGUGGCUAGG-CA-------------------AACUGGCCAA ...((((((.....((..((((.((((((....-......))))))))))))...(((..(((((......-------..))))).))-).-------------------...)))))). ( -28.80) >DroSec_CAF1 93612 99 - 1 ACUUGGCCAAUUAAGCAAUUACGCUAACAAGCU-UUAUUAUGUUGGGUAAGCAAGGCUGAAGCCAACAAUGGGUAAUGGGUGGCCAGG-CA-------------------AACUGGCCAA .((..(((......((..((((.((((((....-......))))))))))))..(((....))).......)))...)).(((((((.-..-------------------..))))))). ( -31.20) >DroSim_CAF1 52539 99 - 1 ACUUGGCCAAUUAAGCAAUUACGCUAACAAGCU-UUAUUAUGUUGGGUAAGCAAGGCUGAAGCCAACAAUGGGUAAUGGGUGGCCAGG-CA-------------------AACUGGCCAA .((..(((......((..((((.((((((....-......))))))))))))..(((....))).......)))...)).(((((((.-..-------------------..))))))). ( -31.20) >DroEre_CAF1 43974 91 - 1 ACUUGGCCAAUUAAGCAAUUACGCUAACAAGCU-UUAUUAUGUUGGGCAAGCAAGGCUGAAGCCAACAAUG-------GGUGGGCAGU-CA--------------------ACUGGCCAA ...((((((.............(((....(((.-.......))).)))......(((((..(((.......-------)))...))))-).--------------------..)))))). ( -25.60) >DroYak_CAF1 49562 90 - 1 ACUUGGCCAAUUAAGCAAUUACGCUAACAAGCU-UUAUUAUGUUGGGUAAGCAAGACUGA-GCCAACAAUG-------GGUGGCUAGU-CA--------------------ACUGGCCAA ...((((((.....((..((((.((((((....-......))))))))))))..((((.(-((((......-------..))))))))-).--------------------..)))))). ( -30.20) >DroAna_CAF1 47983 115 - 1 UCCUGGCCAAUUAAGCAAUUACGCUAACAAGCUUUUAUUAUGUUGUGCAAGCCGGGCUGAAACCAACAAUGGCC-----AUGGCCAAGUCAAUCCCGUGACCCACCCAGGAACUAGCCAA (((((((....(((....))).(((((((...........))))).))..)))(((.((..........((((.-----...)))).((((......)))).)))))))))......... ( -27.00) >consensus ACUUGGCCAAUUAAGCAAUUACGCUAACAAGCU_UUAUUAUGUUGGGUAAGCAAGGCUGAAGCCAACAAUG_______GGUGGCCAGG_CA___________________AACUGGCCAA .((((.((((((((....))).(((....))).........))))).))))...(((....)))................(((((((.........................))))))). (-19.00 = -19.78 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:18 2006