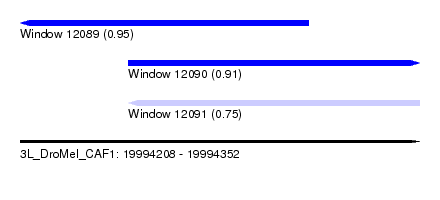
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,994,208 – 19,994,352 |
| Length | 144 |
| Max. P | 0.953149 |

| Location | 19,994,208 – 19,994,312 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -26.67 |
| Energy contribution | -26.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19994208 104 - 23771897 AUAUCUAUAUGUAUACAAUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGUUAAUGGAGUGUGCCGAGGGCAGAACUGCCGCUUUAUACUGCU ((((((((.(((((((.(((((((((........))))))))).))))))).))))))))......(.((((((..(((((((....)))).))))))))).). ( -30.20) >DroSec_CAF1 84082 100 - 1 AUA---GUAUAUAUAC-AUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGCUAAUGGAGUGUGCCGAGGGCAGAACUGCCGCUUUAUACUGCU ...---((((((...(-(((.......))))....))))))(..(((((.((((.((((..((.....))..))))...((((....)))))))))))))..). ( -29.50) >DroSim_CAF1 42812 100 - 1 AUA---GUAUAUAUAC-AUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGUUAAUGGAGUGUGCCGAGGGCAGAACUGCCGCUUUAUACUGCU ...---((((((...(-(((.......))))....))))))(..(((((.((((.((((..((.....))..))))...((((....)))))))))))))..). ( -26.60) >consensus AUA___GUAUAUAUAC_AUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGUUAAUGGAGUGUGCCGAGGGCAGAACUGCCGCUUUAUACUGCU ..........((((((..((((.....))))....))))))(..(((((.((((.((((..((.....))..))))...((((....)))))))))))))..). (-26.67 = -26.23 + -0.44)



| Location | 19,994,247 – 19,994,352 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -11.40 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19994247 105 + 23771897 UUAACACACCCACUUACACACCACAUACACCAUCCAUAAAAUAUAUAUUGUAUACAUAUAGAUAUAUGUACAAACAUCUGAUAUCAAUGACUUUUGUAGCUGUUU .....................((..((((.....(((...((((....(((.(((((((....)))))))...)))....))))..))).....))))..))... ( -8.50) >DroSec_CAF1 84121 101 + 1 UUAGCACACCCACUUACACACCACAUACACCAUCCAUAAAAUAUAUAU-GUAUAUAUAC---UAUAUGUACACUCAACUGAUAUCAAUGACUUGUGUAGCUGUUU .((((((((......................................(-((((((((..---.))))))))).(((..((....)).)))...)))).))))... ( -12.90) >DroSim_CAF1 42851 101 + 1 UUAACACACCCACUUACACACCACAUACACCAUCCAUAAAAUAUAUAU-GUAUAUAUAC---UAUAUGUACACACAACUGAUAUCAAUGACUUGUGUAGCUGUUU ..........((.(((((((...(((.....................(-((((((((..---.)))))))))......((....)))))...))))))).))... ( -12.80) >consensus UUAACACACCCACUUACACACCACAUACACCAUCCAUAAAAUAUAUAU_GUAUAUAUAC___UAUAUGUACACACAACUGAUAUCAAUGACUUGUGUAGCUGUUU ..........((.(((((((...(((.......................(((((((((....))))))))).......((....)))))...))))))).))... (-11.20 = -11.87 + 0.67)

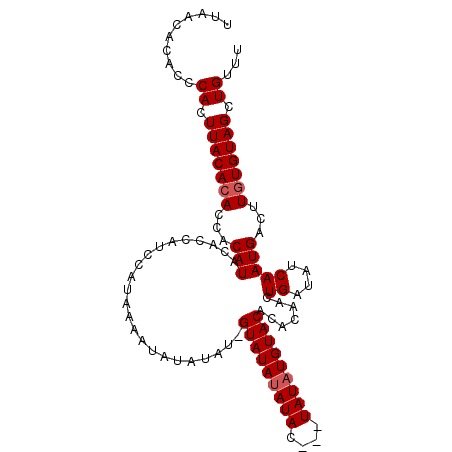
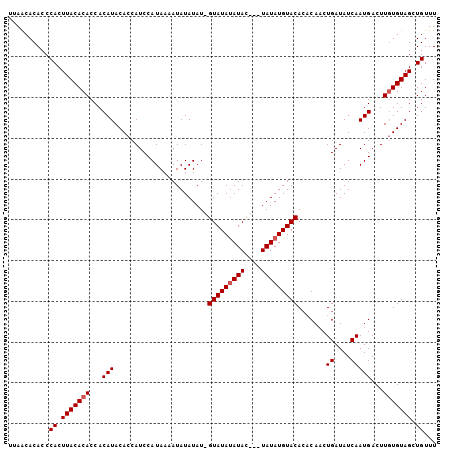
| Location | 19,994,247 – 19,994,352 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -20.02 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19994247 105 - 23771897 AAACAGCUACAAAAGUCAUUGAUAUCAGAUGUUUGUACAUAUAUCUAUAUGUAUACAAUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGUUAA .....(.((((((.(((..((....))))).)))))))((((((((((.(((((((.(((((((((........))))))))).))))))).))))))))))... ( -24.00) >DroSec_CAF1 84121 101 - 1 AAACAGCUACACAAGUCAUUGAUAUCAGUUGAGUGUACAUAUA---GUAUAUAUAC-AUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGCUAA ..(((.(((((((..((((((....))).))).)))..(((((---.(((((((((-.((((.....))))....))))))))).)))))..)))).)))..... ( -25.40) >DroSim_CAF1 42851 101 - 1 AAACAGCUACACAAGUCAUUGAUAUCAGUUGUGUGUACAUAUA---GUAUAUAUAC-AUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGUUAA ..(((.(((((((..((((..((((((..(((((((((.....---)))))))))(-(((.......))))..))))))..))))..)))..)))).)))..... ( -25.20) >consensus AAACAGCUACACAAGUCAUUGAUAUCAGUUGUGUGUACAUAUA___GUAUAUAUAC_AUAUAUAUUUUAUGGAUGGUGUAUGUGGUGUGUAAGUGGGUGUGUUAA ..(((.((((......(((((....))).))....(((((((.....(((((((((..((((.....))))....)))))))))))))))).)))).)))..... (-20.02 = -19.80 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:10 2006