| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,993,122 – 19,993,234 |
| Length | 112 |
| Max. P | 0.768278 |

| Location | 19,993,122 – 19,993,234 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -32.87 |
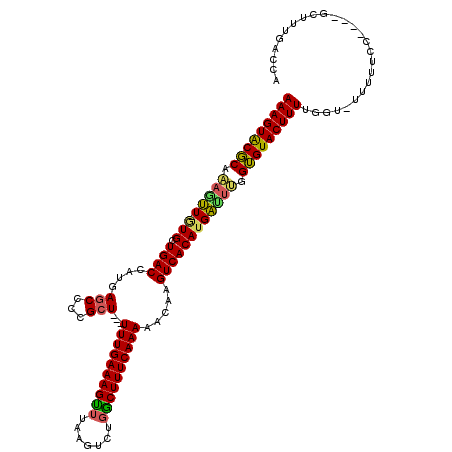
| Consensus MFE | -25.52 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19993122 112 - 23771897 AAAGUACGCAAAGUUAAUGCUGACCAUGAGCCCCGCU---UUUGAAAGUUUAAGUCUGGCUUUCAAAAACAAGUCACAUGAUUUGGUGUACUUUUGGU-UUUUUCC----GCUUUGACCA (((((((((.((((((.((.((((...((((...)))---)(((((((((.......)))))))))......)))))))))))).)))))))))((((-(......----.....))))) ( -29.50) >DroSec_CAF1 83055 111 - 1 AAAGUACGCAAAGUU-GUGCUGACCAUGAGCCCCGCU---UUUGAAAGCUUAAGUCUGGCUUUCAAAAACAAGUCACAUGAUUUGGUGUACUUUUGGU-UGUUUCC----GCUUUGACCA (((((((((.((((.-.((.((((...((((...)))---)(((((((((.......)))))))))......))))))..)))).)))))))))((((-((.....----....)))))) ( -35.10) >DroSim_CAF1 41799 112 - 1 AAAGUACGCAAAGUU-GUGCUGACCAUGAGCCCCGCU---UUUGAAAGCUUAAGUCUGGCUUUCAAAAACAAGUCACAUGAUUUGGUGUACUUUUGGUUUAUUUCC----GCUUUGACCA (((((((((.((((.-.((.((((...((((...)))---)(((((((((.......)))))))))......))))))..)))).)))))))))(((((.......----.....))))) ( -34.80) >DroEre_CAF1 34063 111 - 1 AAAGUACGCAAACUU-GUGCUGACCAAGAGCCCCGCU---UUUGAAAGUUUAAGUCUGGCUUUCAAAAACAAGUCACAUGAUUUGGUGUACUUUUGGU-UUUUUCC----GCUUUGAUCA ..((((((......)-)))))(((((((((...((((---((((((((((.......))))))))))).((((((....)))))))))..))))))))-)......----.......... ( -31.10) >DroYak_CAF1 34806 111 - 1 AAAGUACGCAAAGUU-GUGCUGACCAUGAGCCCCGCU---UUUGAAAGUUUAAGUCUGGCUUUCAAAAACAAGUCACAUGACUGGGUGUACUUUUGGU-UUUUCCC----GAUUUGAUCA .((((((((..(((.-.((.((((...((((...)))---)(((((((((.......)))))))))......))))))..)))..))))))))((((.-.....))----))........ ( -33.40) >DroAna_CAF1 36228 111 - 1 AAAGUGCUCAGAAGU-GUGGUGACCACGAGCCCCGAUUUUUUUGAAAGU-------UGACUUUCAAAGAAAAGUCACAAGCUCCGGGGUACUUUUGGU-UUUUUCGACAAACUUUCACCA (((((...(((((((-((((...))))..((((((.((((((((((((.-------...))))))))))))(((.....))).)))))))))))))((-(.....)))..)))))..... ( -33.30) >consensus AAAGUACGCAAAGUU_GUGCUGACCAUGAGCCCCGCU___UUUGAAAGUUUAAGUCUGGCUUUCAAAAACAAGUCACAUGAUUUGGUGUACUUUUGGU_UUUUUCC____GCUUUGACCA (((((((((.(((((.(((.((((....(((...)))...((((((((((.......)))))))))).....)))))))))))).))))))))).......................... (-25.52 = -24.92 + -0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:06 2006