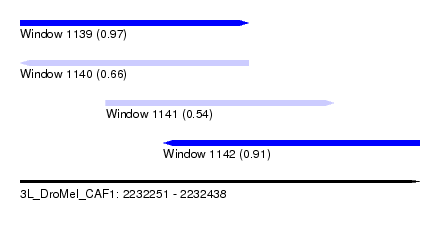
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,232,251 – 2,232,438 |
| Length | 187 |
| Max. P | 0.967534 |

| Location | 2,232,251 – 2,232,358 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2232251 107 + 23771897 UGAAGGUAGGCCAGGUAGACCAUCUUGUGUGGCUUAACCACUAAGAU-------------UGUUACCAAUUUCAGAACAAGUGUCAGAAUCUACGCCUGGAAUUGGCCUGCUCUCGCCUU .((..(((((((.(((.(((.((((((.((((.....))))))))))-------------.))))))((((((((.....((((........)))))))))))))))))))..))..... ( -40.40) >DroSec_CAF1 13187 106 + 1 UGAAGGUAGACCAGG-AUUCCGUCUUGUGUGGCUAACCCACUUAGAU-------------UGUUACCAAUUUCAGAACAAGUGCCAGAAGCUACGCCUGGAAUUGGCCUGCUCUCGCCUU ..((((((((.((((-.....((((...((((.....))))..))))-------------.....((((((((((.....((((.....)).))..)))))))))))))).))).))))) ( -33.30) >DroSim_CAF1 13263 106 + 1 UGAAGGUAGACCAGG-AUUCUGUCUUGUGUGGCUUACCCACAUAGAU-------------UGUUACCAAUUUCAGAACAAGUGUCAGAAUCUUCGCCUGGAAUUGGCCUGCUCUCGCCUU ..((((((((.((((-.....((((.((((((.....))))))))))-------------.....((((((((((...(((.........)))...)))))))))))))).))).))))) ( -34.20) >DroEre_CAF1 26240 117 + 1 UGAAGGUAGACCGGG-AUUCCAUCUG--AUGGCCCACCCACUUCAGUCUCCACCACGCUCUAAUUCCAAUUCCAGAGCAAGUGUCAGAGUCUACGCCUGGAAAUGGCCUGCUCCCACCUC ((..(((((.(((((-...((((...--)))))))..(((....((.(((...((((((((............)))))..)))...))).)).....)))....)).)))))..)).... ( -27.00) >DroYak_CAF1 13714 109 + 1 UGAAGGUAGACCAGG-ACUCCAUCUCGUAUGGCCUACC-ACU--------CACCACU-UCCUAUUCCAAUUACAGGAUAAGUGCCAGAGUCUACGCCUGGAAAUGGCCUGCUCUCACCUC ...(((((((.((((-...((((.((....(((.....-(((--------(..((((-((((...........))))..))))...))))....)))..)).)))))))).))).)))). ( -29.20) >consensus UGAAGGUAGACCAGG_AUUCCAUCUUGUGUGGCUUACCCACUUAGAU_____________UGUUACCAAUUUCAGAACAAGUGUCAGAAUCUACGCCUGGAAUUGGCCUGCUCUCGCCUU ...(((((((.((((.............((((.....))))........................((((((((((.....(((..........))))))))))))))))).))).)))). (-21.50 = -22.22 + 0.72)

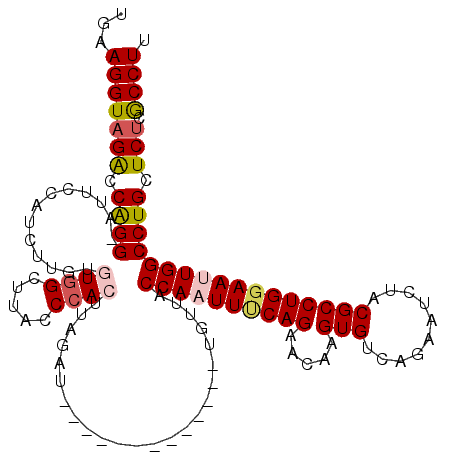
| Location | 2,232,251 – 2,232,358 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2232251 107 - 23771897 AAGGCGAGAGCAGGCCAAUUCCAGGCGUAGAUUCUGACACUUGUUCUGAAAUUGGUAACA-------------AUCUUAGUGGUUAAGCCACACAAGAUGGUCUACCUGGCCUACCUUCA .((((.((.(.((((((....((((.......))))...(((((..(((.((((....))-------------)).)))((((.....))))))))).)))))).))).))))....... ( -31.90) >DroSec_CAF1 13187 106 - 1 AAGGCGAGAGCAGGCCAAUUCCAGGCGUAGCUUCUGGCACUUGUUCUGAAAUUGGUAACA-------------AUCUAAGUGGGUUAGCCACACAAGACGGAAU-CCUGGUCUACCUUCA ((((..(((.((((...(((((.......((.....)).(((((......((((....))-------------))....((((.....)))))))))..)))))-)))).))).)))).. ( -29.70) >DroSim_CAF1 13263 106 - 1 AAGGCGAGAGCAGGCCAAUUCCAGGCGAAGAUUCUGACACUUGUUCUGAAAUUGGUAACA-------------AUCUAUGUGGGUAAGCCACACAAGACAGAAU-CCUGGUCUACCUUCA ((((..(((.((((((.......)))...(((((((....(((((((......)).))))-------------)(((.((((((....)).)))))))))))))-)))).))).)))).. ( -33.20) >DroEre_CAF1 26240 117 - 1 GAGGUGGGAGCAGGCCAUUUCCAGGCGUAGACUCUGACACUUGCUCUGGAAUUGGAAUUAGAGCGUGGUGGAGACUGAAGUGGGUGGGCCAU--CAGAUGGAAU-CCCGGUCUACCUUCA ((((((((....((((...((((....(((.((((..(((..(((((((........))))))))))..)))).)))...))))..))))..--....(((...-.))).)))))))).. ( -40.50) >DroYak_CAF1 13714 109 - 1 GAGGUGAGAGCAGGCCAUUUCCAGGCGUAGACUCUGGCACUUAUCCUGUAAUUGGAAUAGGA-AGUGGUG--------AGU-GGUAGGCCAUACGAGAUGGAGU-CCUGGUCUACCUUCA (((((.(((.(((((((((((..(((.((.((((.(.((((..((((((.......))))))-)))).))--------)))-..)).)))....)))))))...-)))).)))))))).. ( -41.60) >consensus AAGGCGAGAGCAGGCCAAUUCCAGGCGUAGAUUCUGACACUUGUUCUGAAAUUGGUAACA_____________AUCUAAGUGGGUAAGCCACACAAGAUGGAAU_CCUGGUCUACCUUCA ((((..(((.((((((((((.((((...((.........))...)))).))))))........................((((.....)))).............)))).))).)))).. (-20.54 = -20.74 + 0.20)

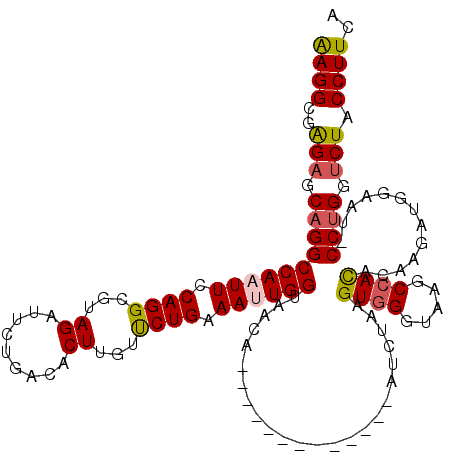
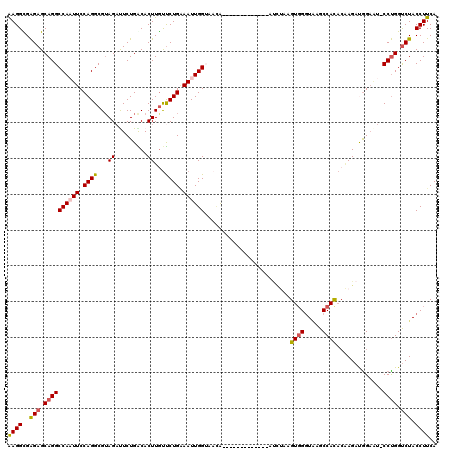
| Location | 2,232,291 – 2,232,398 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2232291 107 + 23771897 CUAAGAU-------------UGUUACCAAUUUCAGAACAAGUGUCAGAAUCUACGCCUGGAAUUGGCCUGCUCUCGCCUUCGUGAGCUGGCCCUGAUGUCGAAUGUGGUUCAAAUUAUUU ....(((-------------(...((((..(((.((......(((((..((((....))))...((((.((((.((....)).)))).))))))))).)))))..))))...)))).... ( -28.30) >DroSec_CAF1 13226 107 + 1 CUUAGAU-------------UGUUACCAAUUUCAGAACAAGUGCCAGAAGCUACGCCUGGAAUUGGCCUGCUCUCGCCUUCGGGAGCUGGCCCUGAUGUCGAAUGUGGUUCAAAUUAUUU ....(((-------------(...((((..(((.((.(((((.((((.........)))).)))((((.(((((((....))))))).))))....)))))))..))))...)))).... ( -31.60) >DroSim_CAF1 13302 107 + 1 CAUAGAU-------------UGUUACCAAUUUCAGAACAAGUGUCAGAAUCUUCGCCUGGAAUUGGCCUGCUCUCGCCUUCGGGAGCUGGCCCUGAUGUCGAAUGUGGUUCAAAUUAUUU ....(((-------------(...((((..(((.((......(((((..(((......)))...((((.(((((((....))))))).))))))))).)))))..))))...)))).... ( -31.40) >DroEre_CAF1 26277 120 + 1 CUUCAGUCUCCACCACGCUCUAAUUCCAAUUCCAGAGCAAGUGUCAGAGUCUACGCCUGGAAAUGGCCUGCUCCCACCUCCGGGAGCUGGCGCUGAUGUCGAAUGUGGUCCAGAUCAUUU .....((((..((((((.((......((.((((((.((..(((........))))))))))).))(((.((((((......)))))).))).........)).))))))..))))..... ( -41.20) >DroYak_CAF1 13752 111 + 1 CU--------CACCACU-UCCUAUUCCAAUUACAGGAUAAGUGCCAGAGUCUACGCCUGGAAAUGGCCUGCUCUCACCUCCGGGAGCUGGCGCUGAUGUCGAAUGUGGUGCAAAUCAUUU ..--------(((((((-((....(((.......)))..(((((((........(((.......)))..((((((......)))))))))))))......))).)))))).......... ( -35.90) >consensus CUUAGAU_____________UGUUACCAAUUUCAGAACAAGUGUCAGAAUCUACGCCUGGAAUUGGCCUGCUCUCGCCUUCGGGAGCUGGCCCUGAUGUCGAAUGUGGUUCAAAUUAUUU ..........................(((((((((.....(((..........))))))))))))(((.((((((......)))))).))).............(((((....))))).. (-22.68 = -23.00 + 0.32)

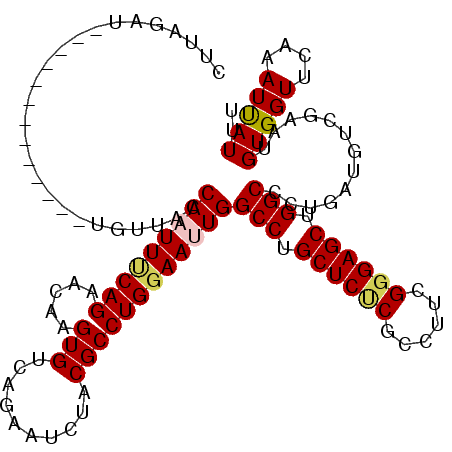
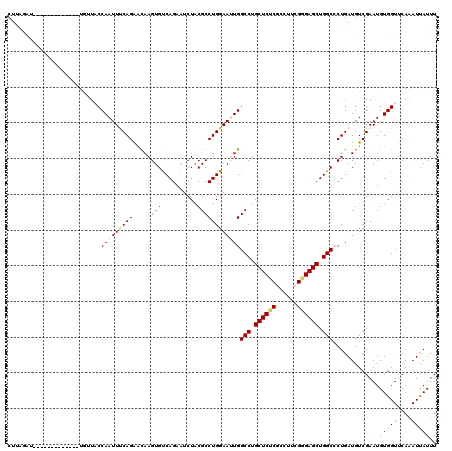
| Location | 2,232,318 – 2,232,438 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -28.80 |
| Energy contribution | -30.28 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2232318 120 - 23771897 AGCAGAAUCACGUGAGCUACCAUCAAUAGUUCCACAUGCCAAAUAAUUUGAACCACAUUCGACAUCAGGGCCAGCUCACGAAGGCGAGAGCAGGCCAAUUCCAGGCGUAGAUUCUGACAC .((((((((..((((((((.......))))).)))(((((.......(((((.....)))))......((((.((((.((....)).)))).)))).......))))).))))))).).. ( -39.60) >DroSec_CAF1 13253 120 - 1 AGCAGAAUCACGUGAGCUACCAUCAAGAGUUCCACAUGCCAAAUAAUUUGAACCACAUUCGACAUCAGGGCCAGCUCCCGAAGGCGAGAGCAGGCCAAUUCCAGGCGUAGCUUCUGGCAC ..(((((.(..(((((((.........)))).)))(((((.......(((((.....)))))......((((.((((.((....)).)))).)))).......))))).).))))).... ( -33.80) >DroSim_CAF1 13329 120 - 1 AGCAGAAUCACGUGAGCUACCAUCAAGAGUUCCACAUGCCAAAUAAUUUGAACCACAUUCGACAUCAGGGCCAGCUCCCGAAGGCGAGAGCAGGCCAAUUCCAGGCGAAGAUUCUGACAC .((((((((..(((((((.........)))).))).((((.......(((((.....)))))......((((.((((.((....)).)))).)))).......))))..))))))).).. ( -37.10) >DroEre_CAF1 26317 120 - 1 AACAGAAUCAUGUGAGCUACCAUCAGUAGUUCCACCUGCCAAAUGAUCUGGACCACAUUCGACAUCAGCGCCAGCUCCCGGAGGUGGGAGCAGGCCAUUUCCAGGCGUAGACUCUGACAC ..((((.((.((.(((((((.....)))))))))...(((...((((.(.((......)).).))))(.(((.(((((((....))))))).)))).......)))...)).)))).... ( -40.60) >DroYak_CAF1 13783 120 - 1 AACAGAAUCACGUGGGAUACCAUCAACAGUUCCACAUGCCAAAUGAUUUGCACCACAUUCGACAUCAGCGCCAGCUCCCGGAGGUGAGAGCAGGCCAUUUCCAGGCGUAGACUCUGGCAC ..((((.((..(((((((..........)))))))((((((((((..((((.((((.((((.....(((....)))..)))).))).).))))..)))))...))))).)).)))).... ( -36.40) >consensus AGCAGAAUCACGUGAGCUACCAUCAAGAGUUCCACAUGCCAAAUAAUUUGAACCACAUUCGACAUCAGGGCCAGCUCCCGAAGGCGAGAGCAGGCCAAUUCCAGGCGUAGAUUCUGACAC ..(((((((..(((((((.........)))).)))(((((.......(((((.....)))))......((((.((((.((....)).)))).)))).......))))).))))))).... (-28.80 = -30.28 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:03 2006