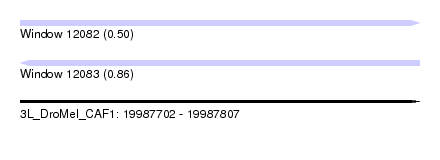
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,987,702 – 19,987,807 |
| Length | 105 |
| Max. P | 0.861555 |

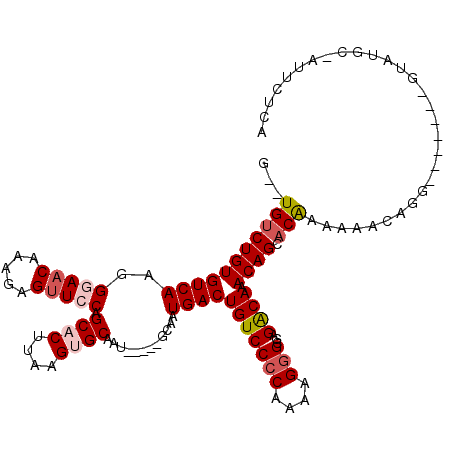
| Location | 19,987,702 – 19,987,807 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19987702 105 + 23771897 UGAGAAU-GCAUAU-------CCUGUUUUUUGUUCUGUUUGUCUCGCCCUUUUGGGGACAGUCAUUGC-----AUUGCACUUAAGUGCUGGAACUCUUUGUUUCCUUGACACAGACA--C ..((((.-(((...-------..))).))))..(((((.((((...(((....)))))))((((....-----...((((....)))).(((((.....)))))..)))))))))..--. ( -24.70) >DroSec_CAF1 77755 105 + 1 UGAGAAU-GAAUAU-------CCUGUUUUUUGUGCUGUUUGUCUCGCCCUUUUGGGGACAGUCAUUGC-----AUUGCACUUAAGUGCUGGAACUCUUUGUUCCCUUGACACAGACA--C .......-......-------..........(((((((.((((...(((....)))))))((((....-----...((((....)))).(((((.....)))))..)))))))).))--) ( -27.90) >DroSim_CAF1 36502 104 + 1 UGAGAAU-GCAUAC-------CCUGUU-UUUGUGCUGUUUGUCUCGCCCUUUUGGGGACAGUCAUUGC-----AUUGCACUUAAGUGCUGGAACUCUUUGUUCCCUUGACACAGACA--C .......-(((((.-------......-..)))))(((((((.((.(((....)))))..((((....-----...((((....)))).(((((.....)))))..)))))))))))--. ( -29.10) >DroEre_CAF1 28260 94 + 1 -----------------------UGUUUUUUGUGCUGUUUGUCUCGC-CUUUUGGGGACAGUCAUUGCAUUGCAUUGCACUUAAGAGCUGCAACUCUUUGUUCCCUUGACACAGACA--C -----------------------............(((((((.(((.-.....(((((((.....((((......))))...(((((......)))))))))))).))).)))))))--. ( -24.10) >DroYak_CAF1 29548 110 + 1 UAAGGAUUGCGCACCA---UUUCUGUUUUUUGUGCUGUUUGCCUCGCCCUUUUGGGGACAGUCAUUGC-----AUUGCACUUAAGAGCUGGAACUCUUUGUUCCCUUGACACAGACA--C ........(((((...---...........)))))((((((..((...((((((((..((((......-----))))..))))))))..(((((.....)))))...))..))))))--. ( -25.04) >DroAna_CAF1 31628 107 + 1 --------GGGCACCCCGGAUGCCCUUUUUCGUCCUGUUUGCCUCGCCCUUUUGGGGACAGUCAUUGC-----AUUGCACUUAAGUGCUGGAGCGCUUUGUUCCCCUGACACAGGCAGGC --------((((((....).)))))........((((((((..((.(((....)))))..((((....-----...((((....)))).(((((.....)))))..)))).)))))))). ( -38.30) >consensus UGAGAAU_GCAUAC_______CCUGUUUUUUGUGCUGUUUGUCUCGCCCUUUUGGGGACAGUCAUUGC_____AUUGCACUUAAGUGCUGGAACUCUUUGUUCCCUUGACACAGACA__C ...............................((.((((.((((((.........))))))((((............((((....)))).(((((.....)))))..)))))))))).... (-20.44 = -20.88 + 0.45)

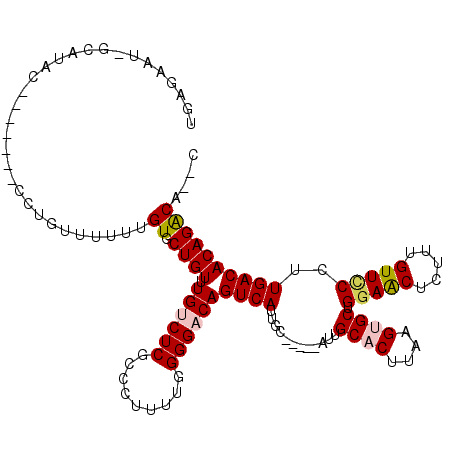
| Location | 19,987,702 – 19,987,807 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19987702 105 - 23771897 G--UGUCUGUGUCAAGGAAACAAAGAGUUCCAGCACUUAAGUGCAAU-----GCAAUGACUGUCCCCAAAAGGGCGAGACAAACAGAACAAAAAACAGG-------AUAUGC-AUUCUCA (--(.(((((((((.((((........)))).((((....))))...-----....))))(((((((....)))...)))).))))))).......(((-------((....-))))).. ( -25.50) >DroSec_CAF1 77755 105 - 1 G--UGUCUGUGUCAAGGGAACAAAGAGUUCCAGCACUUAAGUGCAAU-----GCAAUGACUGUCCCCAAAAGGGCGAGACAAACAGCACAAAAAACAGG-------AUAUUC-AUUCUCA (--((.((((((((..(((((.....))))).((((....))))...-----....))))(((((((....)))...)))).))))))).......(((-------((....-))))).. ( -30.90) >DroSim_CAF1 36502 104 - 1 G--UGUCUGUGUCAAGGGAACAAAGAGUUCCAGCACUUAAGUGCAAU-----GCAAUGACUGUCCCCAAAAGGGCGAGACAAACAGCACAAA-AACAGG-------GUAUGC-AUUCUCA (--((.((((((((..(((((.....))))).((((....))))...-----....))))(((((((....)))...)))).)))))))...-....((-------(.....-...))). ( -31.70) >DroEre_CAF1 28260 94 - 1 G--UGUCUGUGUCAAGGGAACAAAGAGUUGCAGCUCUUAAGUGCAAUGCAAUGCAAUGACUGUCCCCAAAAG-GCGAGACAAACAGCACAAAAAACA----------------------- (--((.((((((((........((((((....))))))...((((......)))).))))(((((((....)-).).)))).)))))))........----------------------- ( -25.60) >DroYak_CAF1 29548 110 - 1 G--UGUCUGUGUCAAGGGAACAAAGAGUUCCAGCUCUUAAGUGCAAU-----GCAAUGACUGUCCCCAAAAGGGCGAGGCAAACAGCACAAAAAACAGAAA---UGGUGCGCAAUCCUUA (--((.((((((((..(((((.....))))).((........))...-----....))))(((((((....)))...)))).)))))))......((....---)).............. ( -27.10) >DroAna_CAF1 31628 107 - 1 GCCUGCCUGUGUCAGGGGAACAAAGCGCUCCAGCACUUAAGUGCAAU-----GCAAUGACUGUCCCCAAAAGGGCGAGGCAAACAGGACGAAAAAGGGCAUCCGGGGUGCCC-------- ((((((((......(((((.......((....((((....))))...-----))........)))))....)))).))))...............((((((.....))))))-------- ( -38.76) >consensus G__UGUCUGUGUCAAGGGAACAAAGAGUUCCAGCACUUAAGUGCAAU_____GCAAUGACUGUCCCCAAAAGGGCGAGACAAACAGCACAAAAAACAGG_______GUAUGC_AUUCUCA ...(((((((((((..(((((.....))))).((((....))))............))))(((((((....)))...)))).)))).))).............................. (-21.08 = -21.88 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:02 2006