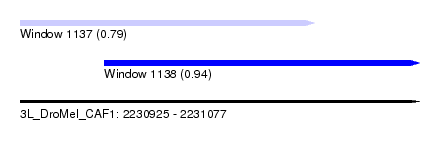
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,230,925 – 2,231,077 |
| Length | 152 |
| Max. P | 0.937421 |

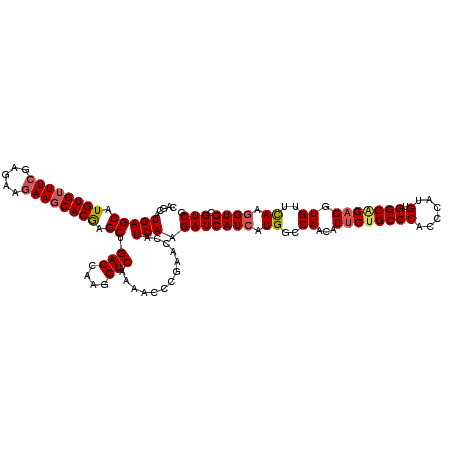
| Location | 2,230,925 – 2,231,037 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2230925 112 + 23771897 GUUUGUUAGUUCAUAUACUUAC--------CUACUUUACUCAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACA (((.((((((((..........--------........(((.....)))...(((((((.....)))))))))))))))...(((((......((....))..(.....).)))))))). ( -25.80) >DroSec_CAF1 11861 112 + 1 GUUCGUUAGUUAACAUACUUAC--------CUACUUUACUCAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACA ((..((.(((......))).))--------..)).........(((((((.((((((((.....)))))))).))((((....)))).....................)))))....... ( -24.10) >DroSim_CAF1 11938 112 + 1 GUUCGUUAGUUCAUAUACUUAC--------CUACUUUACUCAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACA (((.((((((((..........--------........(((.....)))...(((((((.....)))))))))))))))...(((((......((....))..(.....).)))))))). ( -26.10) >DroEre_CAF1 24873 120 + 1 GUUGGUUAGUUCACAUACUUACCCACUCACAUGCUUUUCCCAGCAUGAGCCUGUGCUUCGAAAAGAAACACGAGCAGACCAAGGUCAAAACCCGCACCAUCAAGAUCAUCAUGGCCAACA ((((((((((((.................((((((......)))))).....(((.(((.....))).))))))).(((....))).........................)))))))). ( -30.70) >DroYak_CAF1 12354 116 + 1 GUUGGUUAGUUUACAUACAUACUGAC----CGACUUUUCCUAGCAUGAGCUUGUGUUUCGAAAAAAAGCACAAGCAGACCAAGGUCAAAACCCGCACCAUCAAGAUCAUCAUGGCCAACA ((((((((((..........))))))----)))).........(((((((((((((((.......)))))))))).(((....)))......................)))))....... ( -34.50) >consensus GUUCGUUAGUUCACAUACUUAC________CUACUUUACUCAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACA (((.((..((......)).........................(((((((.((((((((.....)))))))).)).(((....)))......................))))))).))). (-20.54 = -20.78 + 0.24)

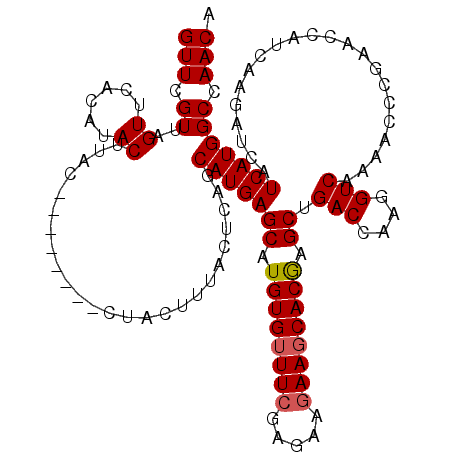
| Location | 2,230,957 – 2,231,077 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -33.48 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2230957 120 + 23771897 CAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACAUUCUGCCCACCCAUGUGGCUGAGGUGUUCAAAGUGCGUCG ..((((.(((.((((((((.....)))))))).)))(((....))).................((.((((.((((((.(((...........))))))))).)))).))...)))).... ( -34.50) >DroSec_CAF1 11893 120 + 1 CAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACAUUCUGCCCACCCAUGUGGCGGAGGUGUUCAAGGUGCGUCG .....(((((.((((((((.....)))))))).))((((....))))............))).(((((((.((..((...(((((((((....)).))))))).))..)).)))).))). ( -38.30) >DroSim_CAF1 11970 120 + 1 CAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACAUUCUGCCCACCCAUGUGGCGGAGGUGUUCAAGGUGCGUCG .....(((((.((((((((.....)))))))).))((((....))))............))).(((((((.((..((...(((((((((....)).))))))).))..)).)))).))). ( -38.30) >DroEre_CAF1 24913 120 + 1 CAGCAUGAGCCUGUGCUUCGAAAAGAAACACGAGCAGACCAAGGUCAAAACCCGCACCAUCAAGAUCAUCAUGGCCAACAUUCUGCCCACCCACGUGGCAGAGGUGUUUAAGGUGCGUCG ........((.((((.(((.....))).)))).)).(((....)))......((((((.....(..(.....)..)(((((((((((.((....)))))))).)))))...))))))... ( -36.20) >DroYak_CAF1 12390 120 + 1 UAGCAUGAGCUUGUGUUUCGAAAAAAAGCACAAGCAGACCAAGGUCAAAACCCGCACCAUCAAGAUCAUCAUGGCCAACAUACUGCCCACCCACGUGGCAGAGGUGUUCAAGGUGCGUCG ........((((((((((.......)))))))))).(((....)))......((((((.....((.(((((((.....))).(((((.((....))))))).)))).))..))))))... ( -39.90) >consensus CAGCAUGAGCAUGUGUUUCGAGAAGAAGCACGAGCUGACCAAGGUCAAAACCCGAACCAUCAAGAUCAUCAUGGCCAACAUUCUGCCCACCCAUGUGGCAGAGGUGUUCAAGGUGCGUCG .....(((((.((((((((.....)))))))).)).(((....))).............))).(((((((.((..((...((((((((......).))))))).))..)).)))).))). (-33.48 = -34.00 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:59 2006