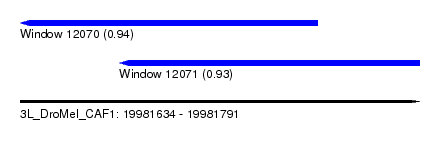
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,981,634 – 19,981,791 |
| Length | 157 |
| Max. P | 0.936596 |

| Location | 19,981,634 – 19,981,751 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -23.23 |
| Energy contribution | -22.73 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19981634 117 - 23771897 AGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCUGCGCAGGAAGUCGGUGG--UCUGUCGGGAAAACAGAAUGGCAGAAU-CGUAAUGGCAAUUU .......((((((...((((((((.((((.((.(((((.(((((((((....)))))))))....)))))..))..--(((((.......))))).)))))))))-.))))))))).... ( -34.60) >DroSec_CAF1 71926 117 - 1 AGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUAGUCAUUCUCCGCGCAGGAAGUCGGUGG--UCUGUCGGGAACACAGAAUGGCAGAAU-CGCAAUGGCAAUUU .......((((((...((((((((.((((.((.(((((.(((((((((....)))))))))....)))))..))..--(((((.(....)))))).)))))))))-.))))))))).... ( -41.30) >DroSim_CAF1 30559 117 - 1 AGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCGUUCUCCGCGCAGGAAGUCGGUGG--UCUGUCGGGAAAACAGAAUGGCAGAAU-CGCAAUGGCAAUUU ...(((((((.......))))))).((((..(.(((((.(((((((((....)))))))))....))))).)((((--(((((((...........)))))).))-)))..))))..... ( -39.50) >DroEre_CAF1 22521 109 - 1 AGAAAAAUGUCAUAAAUGCAUUUUAGCCGGGCAACUUCGGCGGAGAAUUGCCAUUCGCUGGGGAGGAAGUGGGUGG--CCUGUCGGGAAAAGCG--------AAU-CGCAAUGGCAAUUU ...(((((((.......))))))).(((((....))(((((((......((((((((((........)))))))))--)))))))).....(((--------...-)))...)))..... ( -33.10) >DroYak_CAF1 23393 108 - 1 AGAAAAAUGUCAUAAAUGCAUUUUAGACAGGCAACUUCAGCGGGGAAUUGCCAUUCUCUGCGGAGGAAGUC-GUGG--CCAGUCGGGAAAACAG--------AAU-CGCAAUGGCAAUUU .......((((((...((((((((.(((.(((.(((((.(((((((((....)))))))))....))))).-...)--)).)))(......)))--------)))-.))))))))).... ( -33.40) >DroAna_CAF1 25809 111 - 1 AGAAAAAUGUCAUAAAUGUUCUCGGGCUUGGCAGCUUCAA-GGGGAAUUUUUACUCCCCAAACAAGAAGCCAGAAGAACCAGACUGCAAUUCUC--------AAUCCGCAAUGGCAAUUU .......((((((....((((((.(((((((.....))..-(((((........))))).......))))).).))))).....(((.......--------.....))))))))).... ( -25.70) >consensus AGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCCGCGCAGGAAGUCGGUGG__CCUGUCGGGAAAACAG________AAU_CGCAAUGGCAAUUU .......((((((...(((.......(((....(((((.(((((((((....)))))))))....)))))...)))...((((.......)))).............))))))))).... (-23.23 = -22.73 + -0.49)

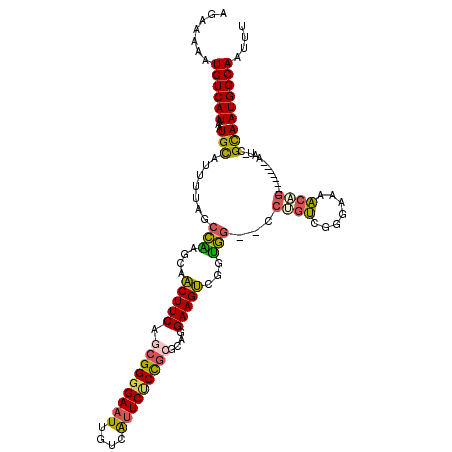
| Location | 19,981,673 – 19,981,791 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -30.58 |
| Energy contribution | -31.75 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19981673 118 - 23771897 GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACUGCAGCUAAGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCUGCGCAGGAAGUCGGUGG--UC ..((((.((((((....)))))).))))((((((..(((..(.(((((((.......)))))))..)..))).(((((.(((((((((....)))))))))....)))))))))))--.. ( -37.10) >DroSec_CAF1 71965 118 - 1 GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACUGCAGCUAAGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUAGUCAUUCUCCGCGCAGGAAGUCGGUGG--UC ..((((.((((((....)))))).))))((((((..(((..(.(((((((.......)))))))..)..))).(((((.(((((((((....)))))))))....)))))))))))--.. ( -39.50) >DroSim_CAF1 30598 118 - 1 GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACUGCAGCUAAGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCGUUCUCCGCGCAGGAAGUCGGUGG--UC ..((((.((((((....)))))).))))((((((..(((..(.(((((((.......)))))))..)..))).(((((.(((((((((....)))))))))....)))))))))))--.. ( -39.60) >DroEre_CAF1 22552 118 - 1 GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACUGCAGCUAAGAAAAAUGUCAUAAAUGCAUUUUAGCCGGGCAACUUCGGCGGAGAAUUGCCAUUCGCUGGGGAGGAAGUGGGUGG--CC ..((((.((((((....)))))).))))((..((((..((.((......)).))..)))).....((((((....))))))))......((((((((((........)))))))))--). ( -36.20) >DroYak_CAF1 23424 117 - 1 GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACUGCAGCUAAGAAAAAUGUCAUAAAUGCAUUUUAGACAGGCAACUUCAGCGGGGAAUUGCCAUUCUCUGCGGAGGAAGUC-GUGG--CC ...(((.((((((....)))))).)))(((((...............((((.((((.....))))))))(((..((((.(((((((((....))))))))).))))..)))-))))--). ( -37.80) >DroAna_CAF1 25841 119 - 1 GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACAGAGGCUAAGAAAAAUGUCAUAAAUGUUCUCGGGCUUGGCAGCUUCAA-GGGGAAUUUUUACUCCCCAAACAAGAAGCCAGAAGAACC ((((((.((((((....))))))....(((.....))).......))))))......((((((.(((((((.....))..-(((((........))))).......))))).).))))). ( -33.80) >consensus GACAUUAAUUGCAUAAGUGCAAUCAAUGCCACUGCAGCUAAGAAAAAUGUCAUAAAUGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCCGCGCAGGAAGUCGGUGG__CC ..((((.((((((....)))))).))))((((((..(((....((((((.((....)))))))).....))).(((((.(((((((((....)))))))))....))))))))))).... (-30.58 = -31.75 + 1.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:51 2006