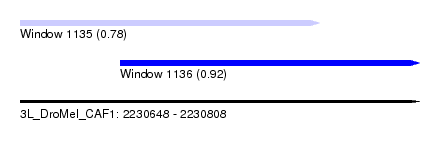
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,230,648 – 2,230,808 |
| Length | 160 |
| Max. P | 0.920311 |

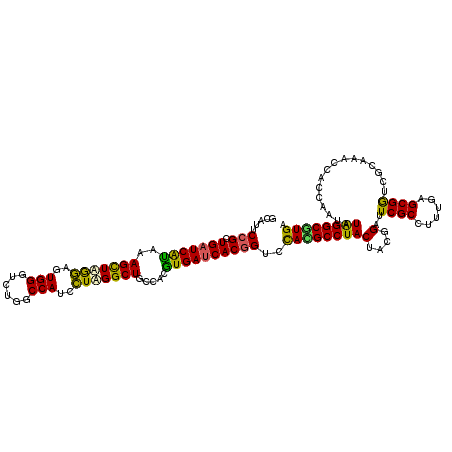
| Location | 2,230,648 – 2,230,768 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -35.32 |
| Energy contribution | -33.72 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2230648 120 + 23771897 GCCGUAGUCAUCGUGCUCAGCCUUAUGUUGGUCAUCAUUGGCAUUCCGCUGAUCAUAAAGUUAGGAGUGGGCCUGGCCAUCCUUGGCUCCCACGUGAUCACUGUACACGCCUACUACGGA .((((((.....(((.(((((...((((..((....))..))))...))))).))).....((((.(((((...(((((....))))))))))(((.........))).)))))))))). ( -44.60) >DroSec_CAF1 11584 120 + 1 GCAGUAAUCAUCGUGCUCAGCCUUAUGUUGGUCAUCAUUGGCAUUCCGCUGAUCAUAAAGCUGGGAGUGGGUCUGGCCAUCCUCGGCUGCCACGUGAUCACGGUCCAUGCCUACUACGGA (.((((......(((..((((.....))))..)))....(((((.(((.(((((((..(((((((..(((......)))..))))))).....))))))))))...))))))))).)... ( -40.90) >DroSim_CAF1 11665 120 + 1 GCAGUAAUCAUCGUGCUCAGCCUCAUGUUGGUCAUCAUUGGCAUUCCGCUGAUCAUAAAGCUGGGAGUGGGUCUGGCCAUCCUCGGCUGCCACGUGAUCACGGUCCACGCCUACUACGGA ...((.((((.((((((((((...((((..((....))..))))...)))))......(((((((..(((......)))..)))))))).)))))))).))(((....)))..(....). ( -38.50) >DroEre_CAF1 24593 120 + 1 GCAGUAGUCAUUGUGCUCAGCCUUUUGCUGAGCAUCAUUGGCAUUCCGCUGAUCGCAAAGCUAGGAGUGGGUCUGGCCAUCCUAGGCUGCCAUGUGACCACGGUCCAUGCCUACUACGGA ...(((((....(((((((((.....)))))))))....(((((.(((.((.(((((.((((((((.(((......)))))))).)))....))))).)))))...))))).)))))... ( -48.80) >DroYak_CAF1 12076 120 + 1 GCAGUAAUCAUUGUGCUUAGCCUUAUGCUGGUCAUCAUUGGCAUUCCGCUGAUCAUAAAGCUAACAGUGGGUCUGGCCAUCGUAGGCUGCCAUAUGAUCACGGUUUACGCCUACUACGGA (.((((......(((.(((((.....))))).)))....(((...(((.((((((((.((((.((.((((......)))).)).))))....))))))))))).....))))))).)... ( -34.50) >consensus GCAGUAAUCAUCGUGCUCAGCCUUAUGUUGGUCAUCAUUGGCAUUCCGCUGAUCAUAAAGCUAGGAGUGGGUCUGGCCAUCCUAGGCUGCCACGUGAUCACGGUCCACGCCUACUACGGA ...((.((((.((((.(((((...((((..((....))..))))...)))))......(((((((..(((......)))..)))))))..)))))))).))........((......)). (-35.32 = -33.72 + -1.60)

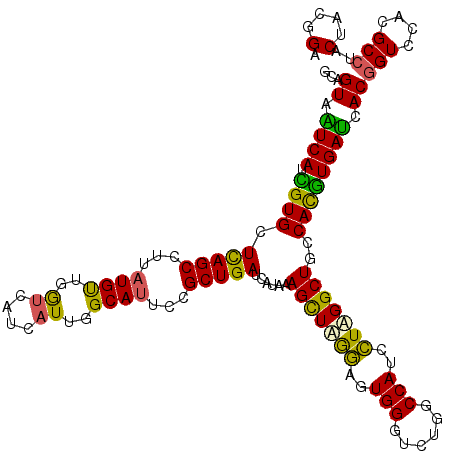
| Location | 2,230,688 – 2,230,808 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -38.00 |
| Energy contribution | -36.80 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2230688 120 + 23771897 GCAUUCCGCUGAUCAUAAAGUUAGGAGUGGGCCUGGCCAUCCUUGGCUCCCACGUGAUCACUGUACACGCCUACUACGGAUUCGCCUUUGAGCGAUCGCAAACCACCAAUAUAGGCGUGA ((.....))(((((((..........(((((...(((((....))))))))))))))))).....(((((((((....)..((((......))))................)))))))). ( -38.80) >DroSec_CAF1 11624 120 + 1 GCAUUCCGCUGAUCAUAAAGCUGGGAGUGGGUCUGGCCAUCCUCGGCUGCCACGUGAUCACGGUCCAUGCCUACUACGGAUUCGCCUUUGAGCGGUCCCAAACCACCGAUAUAGGCGUGU .....(((.(((((((..(((((((..(((......)))..))))))).....))))))))))..((((((((...(((....((......))(((.....))).)))...)))))))). ( -45.90) >DroSim_CAF1 11705 120 + 1 GCAUUCCGCUGAUCAUAAAGCUGGGAGUGGGUCUGGCCAUCCUCGGCUGCCACGUGAUCACGGUCCACGCCUACUACGGAUUCGCCUUUGAGCGGUCCCAAACCACCGAUAUAGGCGUGU .....(((.(((((((..(((((((..(((......)))..))))))).....))))))))))..((((((((...(((....((......))(((.....))).)))...)))))))). ( -47.70) >DroEre_CAF1 24633 120 + 1 GCAUUCCGCUGAUCGCAAAGCUAGGAGUGGGUCUGGCCAUCCUAGGCUGCCAUGUGACCACGGUCCAUGCCUACUACGGAUUCGCCUUCGAGCGGUCGGCAACCACCAAUGUGGGCAUGA ((.....(((((((((....(.(((.((((((((((((......))).(.((((.(((....))))))).).....)))))))))))).).)))))))))..((((....)))))).... ( -47.60) >DroYak_CAF1 12116 120 + 1 GCAUUCCGCUGAUCAUAAAGCUAACAGUGGGUCUGGCCAUCGUAGGCUGCCAUAUGAUCACGGUUUACGCCUACUACGGAUUCGCCUUUGAGCGAUCGGAAACCACCAAUGUGGGCAUGA .....(((.((((((((.((((.((.((((......)))).)).))))....))))))))))).....((((((...((..((((......))))..(....)..))...)))))).... ( -41.50) >consensus GCAUUCCGCUGAUCAUAAAGCUAGGAGUGGGUCUGGCCAUCCUAGGCUGCCACGUGAUCACGGUCCACGCCUACUACGGAUUCGCCUUUGAGCGGUCGCAAACCACCAAUAUAGGCGUGA .....(((.(((((((..(((((((..(((......)))..))))))).....))))))))))..((((((((....((..((((......))))..........))....)))))))). (-38.00 = -36.80 + -1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:57 2006