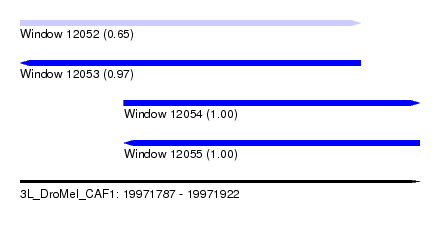
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,971,787 – 19,971,922 |
| Length | 135 |
| Max. P | 0.997583 |

| Location | 19,971,787 – 19,971,902 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -25.18 |
| Energy contribution | -27.46 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19971787 115 + 23771897 GAAAUUCAGACA--GGACUCAGGCGCAGCACGAACAAAGCAAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCGCUAGGACCCUCCUCGUCCUGCGGAGGUCCUUGCCAU ............--.......(((...(((((......((((((.(......).)).)))).....))))).((((((((((.(((((.......))))))))..)))))))))).. ( -36.50) >DroSec_CAF1 62792 96 + 1 GAAAUGCAGACA--GGACACAGGCGCAACACGAACA---------------UUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGAC----CUCGUCCUGCGGAGGUCCUUGCCAU ............--.......(((...(((((....---------------..(((...)))....))))).(((((((..(((((((----...)))))).)..)))))))))).. ( -34.00) >DroSim_CAF1 21301 111 + 1 GAAAUUCAGACA--GGACACAGGCGCAGCACGAACAAAGCGAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGAC----CUCGUCCUGCGGAGGUCCUUGCCAU ............--.......(((...(((((......((((((.(......).)).)))).....))))).(((((((..(((((((----...)))))).)..)))))))))).. ( -37.30) >DroEre_CAF1 13780 101 + 1 A------UGGCAUUGUGCAAAUUC-------AGACAGAGCGAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGACU---UUCGUCCUGGCGAGGUCCUUGCCGC .------..(((..(((((((((.-------((((.(......).))))))))))))))))......(((..((((((((.(((((((.---...))))))).).)))))))..))) ( -42.00) >DroYak_CAF1 13823 106 + 1 G------CGACA--GAGCACAGGCCGAGCACGAACAAAGCGAGCCAUUUAAUUUGCAUUGCAUUACCGUGUGGAGAACCCUCCAGGACC---CUCGUCCUGCUGAGGUCCUUGCCAU (------(....--..))...(((...(((((......((((((.(......).)).)))).....)))))(((....((((((((((.---...))))))..)))))))..))).. ( -30.90) >consensus GAAAU_CAGACA__GGACACAGGCGCAGCACGAACAAAGCGAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGAC____CUCGUCCUGCGGAGGUCCUUGCCAU .....................(((...(((((..(((.(((((........))))).)))......))))).((((((((..((((((.......))))))..).)))))))))).. (-25.18 = -27.46 + 2.28)

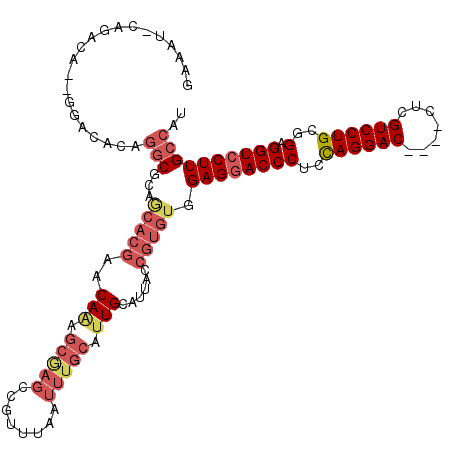
| Location | 19,971,787 – 19,971,902 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -27.84 |
| Energy contribution | -29.60 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19971787 115 - 23771897 AUGGCAAGGACCUCCGCAGGACGAGGAGGGUCCUAGCGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUUGCUUUGUUCGUGCUGCGCCUGAGUCC--UGUCUGAAUUUC ..((((.(((((..(((((.(((((((((..((....))..)))))(((.(((((.((...............))))))).))))))).)))))...).))))--))))........ ( -46.66) >DroSec_CAF1 62792 96 - 1 AUGGCAAGGACCUCCGCAGGACGAG----GUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAA---------------UGUUCGUGUUGCGCCUGUGUCC--UGUCUGCAUUUC ..((((((((((..(.((((((...----)))))))..))))))..((((((...((((((((...---------------.....)))))))).))))))..--))))........ ( -37.60) >DroSim_CAF1 21301 111 - 1 AUGGCAAGGACCUCCGCAGGACGAG----GUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUCGCUUUGUUCGUGCUGCGCCUGUGUCC--UGUCUGAAUUUC ..((((((((((..(.((((((...----)))))))..)))))).((((.(((...(((..(((((.............)))))...)))...)))))))...--))))........ ( -36.72) >DroEre_CAF1 13780 101 - 1 GCGGCAAGGACCUCGCCAGGACGAA---AGUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUCGCUCUGUCU-------GAAUUUGCACAAUGCCA------U (.((..(((((((..(((((((...---.))))))).))))))))).)..((((.((...((((((((..((((......))))..-------.)))))))))).)))).------. ( -45.20) >DroYak_CAF1 13823 106 - 1 AUGGCAAGGACCUCAGCAGGACGAG---GGUCCUGGAGGGUUCUCCACACGGUAAUGCAAUGCAAAUUAAAUGGCUCGCUUUGUUCGUGCUCGGCCUGUGCUC--UGUCG------C ..(((((((((((...((((((...---.))))))..))))))).((((.(((...(((..(((((.............)))))...)))...)))))))...--)))).------. ( -34.92) >consensus AUGGCAAGGACCUCCGCAGGACGAG____GUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUCGCUUUGUUCGUGCUGCGCCUGUGUCC__UGUCUG_AUUUC ..(((((((((((...((((((.......))))))..))))))).((((.(((...(((..(((((.............)))))...)))...))))))).....))))........ (-27.84 = -29.60 + 1.76)

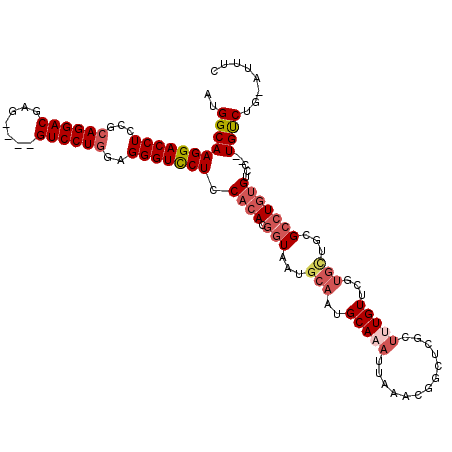
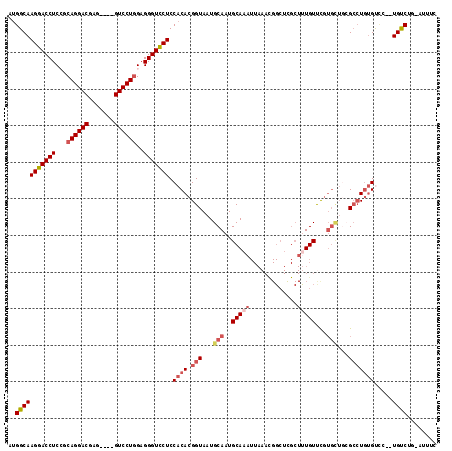
| Location | 19,971,822 – 19,971,922 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.86 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19971822 100 + 23771897 AGCAAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCGCUAGGACCCUCCUCGUCCUGCGGAGGUCCUUGCCAUCGAUAGGUGCAAUUACACUU .(((((........)))))((((((((..((.(((((((((((((.(((((.......))))))))..))))))..))))))...))))))))....... ( -35.00) >DroSec_CAF1 62826 82 + 1 --------------UUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGAC----CUCGUCCUGCGGAGGUCCUUGCCAUCGAUAUGUGCAAUUACACGU --------------.....(((((((...((.((((((((((..(((((((----...)))))).)..))))))..))))))....)))))))....... ( -32.50) >DroSim_CAF1 21336 96 + 1 AGCGAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGAC----CUCGUCCUGCGGAGGUCCUUGCCAUCGAUAGGUGCACUUACACGU .((((((.(......).)).)))).....(((((((((((((..(((((((----...)))))).)..)))))))((.(((....)))))...)))))). ( -36.50) >DroEre_CAF1 13804 93 + 1 AGCGAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGACU---UUCGUCCUGGCGAGGUCCUUGCCGCCGAUAGGUGCAAUUAU---- .(((((........)))))((((((((..((.(((((((((((.(((((((.---...))))))).).))))))..))))))...))))))))...---- ( -41.30) >DroYak_CAF1 13852 97 + 1 AGCGAGCCAUUUAAUUUGCAUUGCAUUACCGUGUGGAGAACCCUCCAGGACC---CUCGUCCUGCUGAGGUCCUUGCCAUCGAUAGGUGCAAUUAUACUU .(((((........)))))((((((((..((.((((.....((((((((((.---...))))))..))))......))))))...))))))))....... ( -28.80) >consensus AGCGAGCCGUUUAAUUUGCAUUGCAUUACCGUGUGGAGGACCCUCCAGGAC____CUCGUCCUGCGGAGGUCCUUGCCAUCGAUAGGUGCAAUUACACGU .(((((........)))))((((((((..((.(((((((((((..((((((.......))))))..).))))))..))))))...))))))))....... (-29.86 = -30.86 + 1.00)


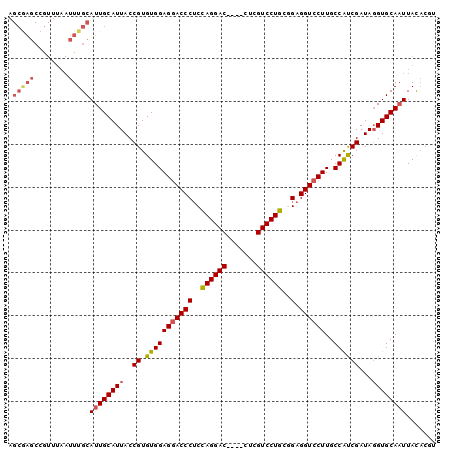
| Location | 19,971,822 – 19,971,922 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -29.70 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19971822 100 - 23771897 AAGUGUAAUUGCACCUAUCGAUGGCAAGGACCUCCGCAGGACGAGGAGGGUCCUAGCGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUUGCU ...((((.(((((..(((((.(((..((((((..((((((((.......))))).))))))))))))..))))).)))))))))................ ( -34.80) >DroSec_CAF1 62826 82 - 1 ACGUGUAAUUGCACAUAUCGAUGGCAAGGACCUCCGCAGGACGAG----GUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAA-------------- ...((((.(((((..(((((.(((..((((((..(.((((((...----)))))))..)))))))))..))))).)))))))))..-------------- ( -34.90) >DroSim_CAF1 21336 96 - 1 ACGUGUAAGUGCACCUAUCGAUGGCAAGGACCUCCGCAGGACGAG----GUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUCGCU ...((((..((((..(((((.(((..((((((..(.((((((...----)))))))..)))))))))..))))).)))).))))................ ( -34.00) >DroEre_CAF1 13804 93 - 1 ----AUAAUUGCACCUAUCGGCGGCAAGGACCUCGCCAGGACGAA---AGUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUCGCU ----...((((((..((((((.((..(((((((..(((((((...---.))))))).))))))))).).))))).))))))................... ( -41.40) >DroYak_CAF1 13852 97 - 1 AAGUAUAAUUGCACCUAUCGAUGGCAAGGACCUCAGCAGGACGAG---GGUCCUGGAGGGUUCUCCACACGGUAAUGCAAUGCAAAUUAAAUGGCUCGCU .......((((((..(((((.(((..(((((((...((((((...---.))))))..))))))))))..))))).))))))................... ( -30.30) >consensus AAGUGUAAUUGCACCUAUCGAUGGCAAGGACCUCCGCAGGACGAG____GUCCUGGAGGGUCCUCCACACGGUAAUGCAAUGCAAAUUAAACGGCUCGCU .......((((((..(((((.(((..(((((((...((((((.......))))))..))))))))))..))))).))))))................... (-29.70 = -30.14 + 0.44)

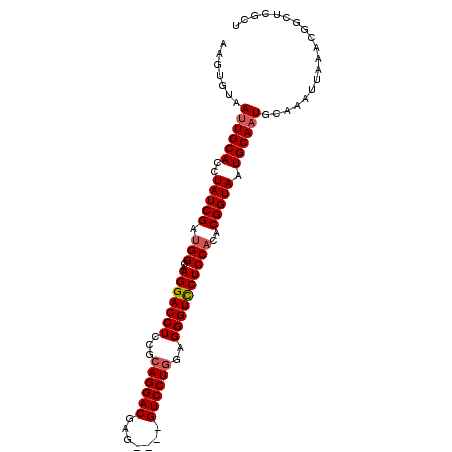
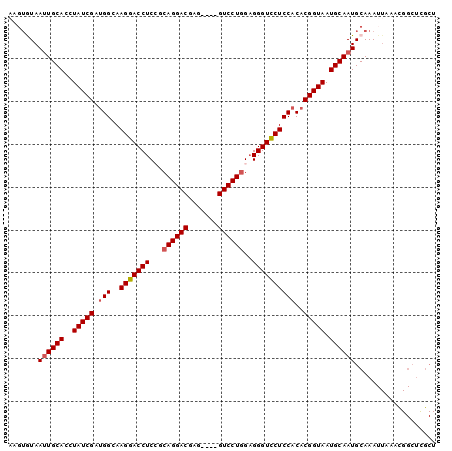
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:35 2006