
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,970,531 – 19,970,662 |
| Length | 131 |
| Max. P | 0.991045 |

| Location | 19,970,531 – 19,970,622 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
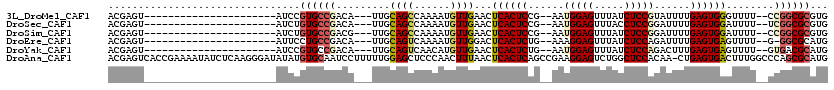
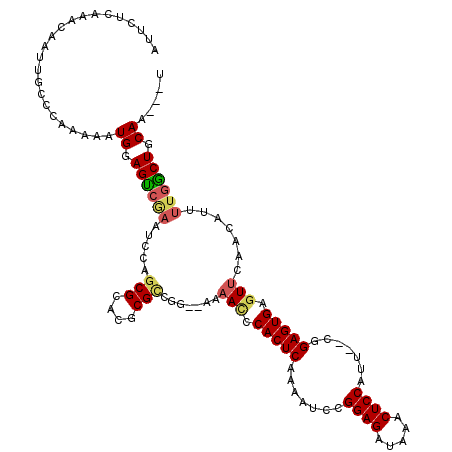
>3L_DroMel_CAF1 19970531 91 + 23771897 ACGAGU----------------------AUCCGUGCCGACA---UUGCAGCCAAAAUGUUGAACUCACUCCG--AAUGGAGUUUAUCUCCGUAUUUUGAGUGGGUUUU--CCGGCGCGUG ......----------------------...((((((((((---((........))))).((((((((((.(--(((((((.....))))))..)).)))))))))).--.))))))).. ( -34.70) >DroSec_CAF1 61478 91 + 1 ACGAGU----------------------AUCUGUGCCGACA---UUGCAGCCAAAAUGUUGAACUCACUCCG--AAUGGAGUUUACCUCCGGAUUUUGAGUGGAUUUU--UCGGCGCGUG ......----------------------....(((((((((---((........))))..(((..(((((.(--(((((((.....))))..)))).)))))..))).--)))))))... ( -27.50) >DroSim_CAF1 20093 91 + 1 ACGAGU----------------------AUCUGUGCCGACG---UUGCAGCCAAAAUGUUGAACUCACUCCG--AAUGGAGUUUAUCUCCGGAUUUUGAGUGGAUUUU--CCGGCGCGUG ......----------------------....((((((..(---((.((((......)))))))((((((.(--(((((((.....))))..)))).)))))).....--.))))))... ( -28.60) >DroEre_CAF1 12547 90 + 1 ACGAGU----------------------AUUCCUGCCGACA---UUGCAGUCAAAAUGUUGGACUCACUCUG--AAAGGAGUUUAUCUCCAGAUUUUGAGUGAGUUUU--G-GGCGCAUG ....((----------------------...(((.((((((---((........))))))))((((((((.(--((.((((.....))))...))).))))))))...--)-)).))... ( -28.50) >DroYak_CAF1 12513 91 + 1 ACGAGU----------------------AUCCGUGCCGACA---UUGCAGUCAACAUGUUGAACUCACUCUG--AAUGGAGUUUAUCUCCAGACUUUGAGUGAGUUUU--GUGACGCAUG ......----------------------...(((((.(((.---.....)))..(((...((((((((((.(--..(((((.....)))))..)...)))))))))).--)))..))))) ( -30.80) >DroAna_CAF1 12201 119 + 1 ACGAGUCACCGAAAAUAUCUCAAGGGAUAUAUGUGCAAUCCUUUUUGGAGCUCCCAACUUUAACUCACUCAGCCGAAGGAGUCUGGCUCCACAA-CUGAGUGACUUUGGCCCAGCGCAUG .((......))...((((((....))))))((((((..(((.....)))....((((.......((((((((.....((((.....))))....-))))))))..))))....)))))). ( -33.20) >consensus ACGAGU______________________AUCCGUGCCGACA___UUGCAGCCAAAAUGUUGAACUCACUCCG__AAUGGAGUUUAUCUCCAGAUUUUGAGUGAGUUUU__CCGGCGCAUG ................................((((((.........((((......))))...((((((......(((((.....)))))......))))))........))))))... (-19.88 = -20.35 + 0.47)
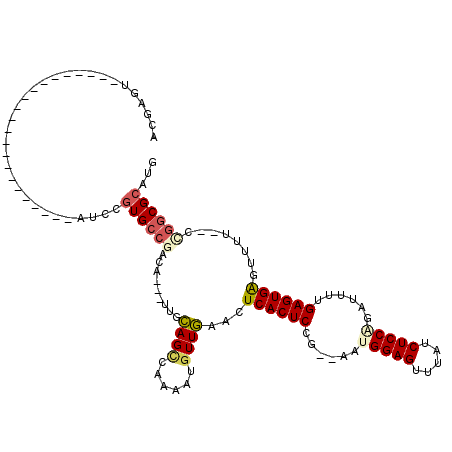

| Location | 19,970,531 – 19,970,622 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19970531 91 - 23771897 CACGCGCCGG--AAAACCCACUCAAAAUACGGAGAUAAACUCCAUU--CGGAGUGAGUUCAACAUUUUGGCUGCAA---UGUCGGCACGGAU----------------------ACUCGU ..((.(((((--......((..((((((...(((....(((((...--.)))))...)))...))))))..))...---..))))).))...----------------------...... ( -25.80) >DroSec_CAF1 61478 91 - 1 CACGCGCCGA--AAAAUCCACUCAAAAUCCGGAGGUAAACUCCAUU--CGGAGUGAGUUCAACAUUUUGGCUGCAA---UGUCGGCACAGAU----------------------ACUCGU ...(((((((--((.....(((((...((((((((......)).))--)))).)))))......))))))).))..---((((......)))----------------------)..... ( -28.10) >DroSim_CAF1 20093 91 - 1 CACGCGCCGG--AAAAUCCACUCAAAAUCCGGAGAUAAACUCCAUU--CGGAGUGAGUUCAACAUUUUGGCUGCAA---CGUCGGCACAGAU----------------------ACUCGU ...(((((((--((.....(((((...((((((...........))--)))).)))))......))))))).))..---.(((......)))----------------------...... ( -23.70) >DroEre_CAF1 12547 90 - 1 CAUGCGCC-C--AAAACUCACUCAAAAUCUGGAGAUAAACUCCUUU--CAGAGUGAGUCCAACAUUUUGACUGCAA---UGUCGGCAGGAAU----------------------ACUCGU .....(((-.--...((((((((..((...((((.....)))).))--..))))))))..........(((.....---.))))))......----------------------...... ( -24.10) >DroYak_CAF1 12513 91 - 1 CAUGCGUCAC--AAAACUCACUCAAAGUCUGGAGAUAAACUCCAUU--CAGAGUGAGUUCAACAUGUUGACUGCAA---UGUCGGCACGGAU----------------------ACUCGU .....(((..--..(((((((((...(..(((((.....)))))..--).)))))))))...(.(((((((.....---.))))))).))))----------------------...... ( -28.00) >DroAna_CAF1 12201 119 - 1 CAUGCGCUGGGCCAAAGUCACUCAG-UUGUGGAGCCAGACUCCUUCGGCUGAGUGAGUUAAAGUUGGGAGCUCCAAAAAGGAUUGCACAUAUAUCCCUUGAGAUAUUUUCGGUGACUCGU ....(((((((...((.((((((((-(((.((((.....))))..))))))))))).))......(((((((((.....)))..)).......))))..........)))))))...... ( -36.81) >consensus CACGCGCCGG__AAAACCCACUCAAAAUCCGGAGAUAAACUCCAUU__CGGAGUGAGUUCAACAUUUUGGCUGCAA___UGUCGGCACAGAU______________________ACUCGU .....(((......(((.(((((.......((((.....)))).......))))).))).........(((.........)))))).................................. (-16.66 = -16.57 + -0.08)

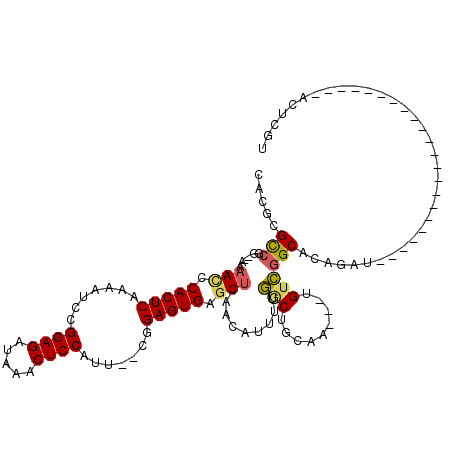
| Location | 19,970,549 – 19,970,662 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19970549 113 + 23771897 A---UUGCAGCCAAAAUGUUGAACUCACUCCG--AAUGGAGUUUAUCUCCGUAUUUUGAGUGGGUUUU--CCGGCGCGUGCGCUGGAUUCGACUCCAUUUUUGGGCAAUUGUUUGAAAAU (---((((..((((((.(((((((((((((.(--(((((((.....))))))..)).))))))))..(--((((((....))))))).))))).....)))))))))))........... ( -41.80) >DroSec_CAF1 61496 113 + 1 A---UUGCAGCCAAAAUGUUGAACUCACUCCG--AAUGGAGUUUACCUCCGGAUUUUGAGUGGAUUUU--UCGGCGCGUGCGCUGGAUUCGACUCCAUUUUUGAGCAAUUGUUUGAAAAU .---.............((((((.((((((.(--(((((((.....))))..)))).))))))....(--((((((....)))))))))))))...(((((..(((....)))..))))) ( -32.90) >DroSim_CAF1 20111 113 + 1 G---UUGCAGCCAAAAUGUUGAACUCACUCCG--AAUGGAGUUUAUCUCCGGAUUUUGAGUGGAUUUU--CCGGCGCGUGCGCUGGAUUCGACUCCAUUUUUGGGCAAUUGUUUGAGAAU (---((((..((((((.((((((.((((((.(--(((((((.....))))..)))).))))))....(--((((((....))))))))))))).....)))))))))))........... ( -38.70) >DroEre_CAF1 12565 112 + 1 A---UUGCAGUCAAAAUGUUGGACUCACUCUG--AAAGGAGUUUAUCUCCAGAUUUUGAGUGAGUUUU--G-GGCGCAUGCGCUGGAUUUGACUCCAUUUUUGGGCAGUUGUUUGAGAAU .---.((.(((((((......(((((((((.(--((.((((.....))))...))).)))))))))((--.-((((....)))).))))))))).))((((..(((....)))..)))). ( -38.10) >DroYak_CAF1 12531 113 + 1 A---UUGCAGUCAACAUGUUGAACUCACUCUG--AAUGGAGUUUAUCUCCAGACUUUGAGUGAGUUUU--GUGACGCAUGCGCUAGAUUUGACUCCAUUUUUGGGCAAUUGUUUGAAAAU .---.(((.((((.((....((((((((((.(--..(((((.....)))))..)...)))))))))))--))))))))(((.(((((..((....))..))))))))............. ( -36.30) >DroAna_CAF1 12241 117 + 1 CUUUUUGGAGCUCCCAACUUUAACUCACUCAGCCGAAGGAGUCUGGCUCCACAA-CUGAGUGACUUUGGCCCAGCGCAUGCGUUUAUUUCGGCUCCAGAUCCGAA--GCCUCACCAGCAU ...(((((((((.((((.......((((((((.....((((.....))))....-))))))))..))))...((((....))))......)))))))))......--((.......)).. ( -33.10) >consensus A___UUGCAGCCAAAAUGUUGAACUCACUCCG__AAUGGAGUUUAUCUCCAGAUUUUGAGUGAGUUUU__CCGGCGCAUGCGCUGGAUUCGACUCCAUUUUUGGGCAAUUGUUUGAAAAU .................((((((.((((((......(((((.....)))))......))))))........(((((....)))))..))))))...(((((..(((....)))..))))) (-23.64 = -24.25 + 0.61)



| Location | 19,970,549 – 19,970,662 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.81 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19970549 113 - 23771897 AUUUUCAAACAAUUGCCCAAAAAUGGAGUCGAAUCCAGCGCACGCGCCGG--AAAACCCACUCAAAAUACGGAGAUAAACUCCAUU--CGGAGUGAGUUCAACAUUUUGGCUGCAA---U ...........((((((((....)))((((((((((.(((....))).))--).(((.(((((..(((..((((.....)))))))--..))))).)))......)))))))))))---) ( -32.40) >DroSec_CAF1 61496 113 - 1 AUUUUCAAACAAUUGCUCAAAAAUGGAGUCGAAUCCAGCGCACGCGCCGA--AAAAUCCACUCAAAAUCCGGAGGUAAACUCCAUU--CGGAGUGAGUUCAACAUUUUGGCUGCAA---U ...........(((((.......((((......))))((....))(((((--((.....(((((...((((((((......)).))--)))).)))))......))))))).))))---) ( -33.00) >DroSim_CAF1 20111 113 - 1 AUUCUCAAACAAUUGCCCAAAAAUGGAGUCGAAUCCAGCGCACGCGCCGG--AAAAUCCACUCAAAAUCCGGAGAUAAACUCCAUU--CGGAGUGAGUUCAACAUUUUGGCUGCAA---C ............((((((((((..(((......(((.(((....))).))--)...)))(((((...((((((...........))--)))).)))))......))))))..))))---. ( -31.30) >DroEre_CAF1 12565 112 - 1 AUUCUCAAACAACUGCCCAAAAAUGGAGUCAAAUCCAGCGCAUGCGCC-C--AAAACUCACUCAAAAUCUGGAGAUAAACUCCUUU--CAGAGUGAGUCCAACAUUUUGACUGCAA---U .............((((((....)))(((((((....(((....))).-.--...((((((((..((...((((.....)))).))--..)))))))).......)))))))))).---. ( -30.10) >DroYak_CAF1 12531 113 - 1 AUUUUCAAACAAUUGCCCAAAAAUGGAGUCAAAUCUAGCGCAUGCGUCAC--AAAACUCACUCAAAGUCUGGAGAUAAACUCCAUU--CAGAGUGAGUUCAACAUGUUGACUGCAA---U ...........((((((((....)))((((((.....(((....)))...--..(((((((((...(..(((((.....)))))..--).))))))))).......))))))))))---) ( -31.00) >DroAna_CAF1 12241 117 - 1 AUGCUGGUGAGGC--UUCGGAUCUGGAGCCGAAAUAAACGCAUGCGCUGGGCCAAAGUCACUCAG-UUGUGGAGCCAGACUCCUUCGGCUGAGUGAGUUAAAGUUGGGAGCUCCAAAAAG ((((..((..(((--((((....)))))))...))....))))((.((.(((..((.((((((((-(((.((((.....))))..))))))))))).))...))).)).))......... ( -43.70) >consensus AUUCUCAAACAAUUGCCCAAAAAUGGAGUCGAAUCCAGCGCACGCGCCGG__AAAACCCACUCAAAAUCCGGAGAUAAACUCCAUU__CGGAGUGAGUUCAACAUUUUGGCUGCAA___U .......................((.((((((.....(((....))).......(((.(((((.......((((.....)))).......))))).))).......)))))).))..... (-21.61 = -21.81 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:30 2006