
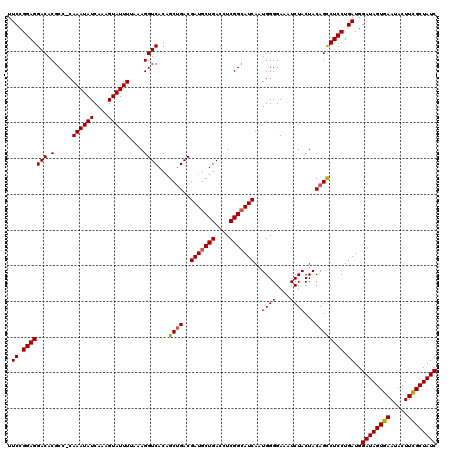
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,943,240 – 19,943,439 |
| Length | 199 |
| Max. P | 0.985200 |

| Location | 19,943,240 – 19,943,359 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.93 |
| Mean single sequence MFE | -33.99 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19943240 119 + 23771897 UUCCGGAGGACACGCC-CAAAUAUCAAAGUAUUUUAAAGGUCACAGCUGACGAUGCUGACCUCGGCAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUUGCUAUC .((.(((((.(...((-((....................((((....))))(((((((....)))))))..))))(....).......)))))).))..(((((..(.....)..))))) ( -33.70) >DroSec_CAF1 8646 120 + 1 UUCCGGAGGACACGCCCCAAAUAUCAAAGUAUUUUAAAGGUCACAGCUGACGAUGCUGACCUCGGAAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUC .((.(((((.....((((((((((....)))))((..((((((..((.......))))))))..)).....))))).............))))).))..((((((((.....)))))))) ( -33.37) >DroSim_CAF1 8193 119 + 1 UUCCGGAGGACACGCC-CAAAUAUCAAAGUAUUUUAAAGGUCACAGCUGACGAUGCUGACCUCGGCAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUC .((.(((((.(...((-((....................((((....))))(((((((....)))))))..))))(....).......)))))).))..((((((((.....)))))))) ( -36.10) >DroYak_CAF1 8777 119 + 1 UUCCGGAGGACACGCC-CAAAUAUCAAAGUAUUUUAAAGGUCACCGCAGACGAUGCUGACCUCGGCAUCAAUGGGGACAUCUACUACAGUCUCCUGAUGGAUAGUGAAUACUUUGCUAUC ....((.(....)..)-)..(((.((((((((((...((((((..(((.....)))))))))...(((((..((((((..........)))))))))))......)))))))))).))). ( -32.80) >consensus UUCCGGAGGACACGCC_CAAAUAUCAAAGUAUUUUAAAGGUCACAGCUGACGAUGCUGACCUCGGCAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUC .((.(((((((.(.....((((((....))))))....))))...((((..(((((((....)))))))..((((....))))...)))))))).))..((((((((.....)))))))) (-30.86 = -30.92 + 0.06)


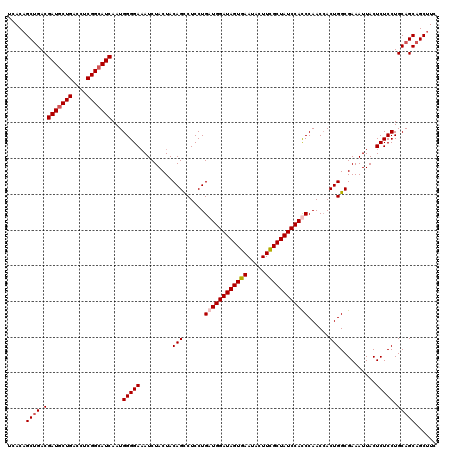
| Location | 19,943,279 – 19,943,399 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -36.41 |
| Energy contribution | -37.16 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19943279 120 + 23771897 UCACAGCUGACGAUGCUGACCUCGGCAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUUGCUAUCCAUCCAACAACUGGCGAAAUUACUCUCCUGCAGCAGCUUC ....(((((.((((((((....)))))))...(((((...........(((....(((((((((..(.....)..)))))))))........)))........)))))....)))))).. ( -40.81) >DroSec_CAF1 8686 120 + 1 UCACAGCUGACGAUGCUGACCUCGGAAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUCCACCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUC ....(((((.((((.(((....))).)))..((((...........(((....))).((((((((((.....))))))))))))))......((.((......)).))....)))))).. ( -33.50) >DroSim_CAF1 8232 120 + 1 UCACAGCUGACGAUGCUGACCUCGGCAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUCCACCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUC ....(((((.((((((((....)))))))..((((...........(((....))).((((((((((.....))))))))))))))......((.((......)).))....)))))).. ( -40.10) >DroYak_CAF1 8816 120 + 1 UCACCGCAGACGAUGCUGACCUCGGCAUCAAUGGGGACAUCUACUACAGUCUCCUGAUGGAUAGUGAAUACUUUGCUAUCCAUCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUC .....((((..(((((((....)))))))...((((((..........)))))).(((((((((..(.....)..))))))))).......................))))......... ( -39.90) >consensus UCACAGCUGACGAUGCUGACCUCGGCAUCAAUGGGGAAAUCUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUCCACCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUC .....((((.((((((((....)))))))...(((((.........(((......((((((((((((.....))))))))))))......)))..........))))).)))))...... (-36.41 = -37.16 + 0.75)


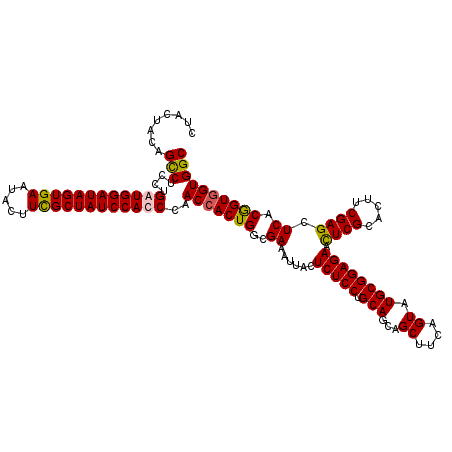
| Location | 19,943,319 – 19,943,439 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -40.04 |
| Energy contribution | -39.98 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19943319 120 + 23771897 CUACUACAGCCUCCUGAUGGAUAGUGAAUACUUUGCUAUCCAUCCAACAACUGGCGAAAUUACUCUCCUGCAGCAGCUUCAGUAUGCGGAGAACUCGCACUUCGAGCUCACGGUGGUGGC (((((((.(((....(((((((((..(.....)..)))))))))........)))((......(((((.(((...((....)).)))))))).((((.....)))).))...))))))). ( -40.30) >DroSec_CAF1 8726 120 + 1 CUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUCCACCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUCAGUAUGCGGAGAACUCGCACUUCGAGCUCACGGUGGUGGC ........(((......((((((((((.....))))))))))....(((((((..((......(((((.(((...((....)).)))))))).((((.....)))).)).)))))))))) ( -41.50) >DroSim_CAF1 8272 120 + 1 CUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUCCACCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUCAGUAUGCGGAGAACUCGCACUUCGAGCUCACGGUGGUGGC ........(((......((((((((((.....))))))))))....(((((((..((......(((((.(((...((....)).)))))))).((((.....)))).)).)))))))))) ( -41.50) >DroYak_CAF1 8856 120 + 1 CUACUACAGUCUCCUGAUGGAUAGUGAAUACUUUGCUAUCCAUCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUCAGUAUGCGGAGAAUUCGCACUUCGAGCUCACAGUGGUGGC ........(((....(((((((((..(.....)..)))))))))..((((((((((((.....(((((.(((...((....)).)))))))).))))).....(....).)))))))))) ( -41.50) >consensus CUACUACAGCCUCCUGAUGGAUAGUGAAUACUUCGCUAUCCACCCAACCACUGGCGAAAUUACUCUCCUGCAGCAGCUUCAGUAUGCGGAGAACUCGCACUUCGAGCUCACGGUGGUGGC ........(((....((((((((((((.....))))))))))))..(((((((..((......(((((.(((...((....)).)))))))).((((.....)))).)).)))))))))) (-40.04 = -39.98 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:20 2006