
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,933,810 – 19,933,940 |
| Length | 130 |
| Max. P | 0.996573 |

| Location | 19,933,810 – 19,933,920 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -22.93 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19933810 110 + 23771897 GAUUUGGGGCCAGGUAUCGAUUUGGUUUUCACUACAGGCUCCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAUAGACCUA----CAAGGAUACCCAAUCA ((((.((((((..(((..((........))..))).)))((((((((.((......(((.(((((((....))))))).)))......))))..----))))))..))))))). ( -38.70) >DroVir_CAF1 3352 107 + 1 GAUUUUGGUCCAGGUAUUGUCUUUGUACGCACAAUUGGUUCCUUGGGCUCCCAAUUUGUAGAAUAAGCUUUCAUAC------AAUUUACACCUAAAAUUAACAUUAGCCAGUC- ((((((((((((((.((((...((((....)))).)))).)).))))).......((((((((......))).)))------))........)))))))..............- ( -16.10) >DroSec_CAF1 1715 110 + 1 GAUUUGGGGCCAGGUAUCGAUUUGGUUUUCACCACAGGCUCCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAAAGACCUA----UAAGGAUACCCAGUCA ((((.((((((((((....))))))).............((((((((.((......(((.(((((((....))))))).)))......))))))----..))))..))))))). ( -37.60) >DroSim_CAF1 1700 110 + 1 GAUUUGGGGCCAGGUAUCGAUUUGGUUUUCACCACAGGCUCCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAUAGACCUA----UAAGGAUAGCCAGUCA ((((((..(((((((....)))))).)..)).....(((((((((((.((......(((.(((((((....))))))).)))......))))))----..))))..))))))). ( -37.90) >DroEre_CAF1 1698 114 + 1 GAUUUGGGGCCAGGUAUUGAUUUAGUUUUCACCACAGGUUCCUUGGGCUCAAUGCUGGUGGAGUACCUCUUGGUACUCAACCUAUAUAGACCUACAUAUAACGAUACCCUUUCA .....((((((..((..(((........)))..)).)))))).((((.((.(((..(((.(((((((....))))))).)))..))).)))))).................... ( -32.20) >DroYak_CAF1 1702 114 + 1 GACUUGGGGCCAGGUAUUGAUUUAGUUUUCACCACAGGUUCCUUGGGCUCAAUGUUGGUGGAGUACCUCUUGGUACUCAACCUAUAUAGACCUACAUAUAACGAAACCCCAUCA ....(((((((..((..(((........)))..)).)).....((((.((.((((.(((.(((((((....))))))).))).)))).))))))............)))))... ( -33.80) >consensus GAUUUGGGGCCAGGUAUCGAUUUGGUUUUCACCACAGGCUCCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAUAGACCUA____UAACGAUACCCAGUCA .....((((((.(((...((........)))))...)))))).((((.((......(((.(((((((....))))))).)))......)))))).................... (-22.93 = -23.77 + 0.84)

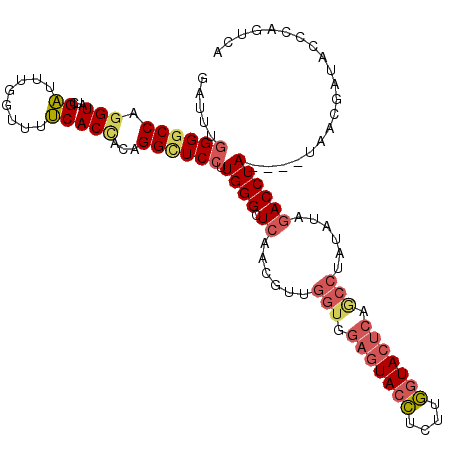
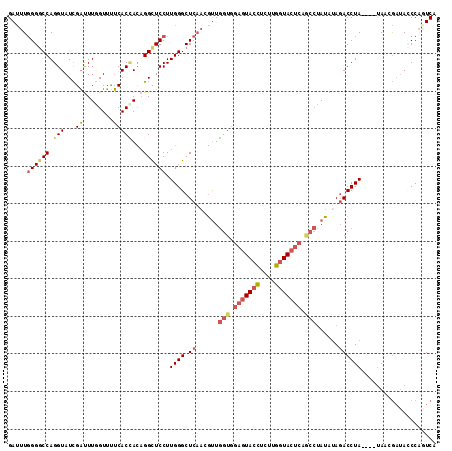
| Location | 19,933,810 – 19,933,920 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19933810 110 - 23771897 UGAUUGGGUAUCCUUG----UAGGUCUAUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGGAGCCUGUAGUGAAAACCAAAUCGAUACCUGGCCCCAAAUC ...(((((..((((((----..((..((....((..(((((((....)))))))..)).....))..))))))))(((.(((.(((........))).)))..))))))))... ( -35.90) >DroVir_CAF1 3352 107 - 1 -GACUGGCUAAUGUUAAUUUUAGGUGUAAAUU------GUAUGAAAGCUUAUUCUACAAAUUGGGAGCCCAAGGAACCAAUUGUGCGUACAAAGACAAUACCUGGACCAAAAUC -...((((....))))..(((((((((...((------(((((...........(((((.((((...((...))..)))))))))))))))).....)))))))))........ ( -20.20) >DroSec_CAF1 1715 110 - 1 UGACUGGGUAUCCUUA----UAGGUCUUUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGGAGCCUGUGGUGAAAACCAAAUCGAUACCUGGCCCCAAAUC ...(..((((((...(----(((((((((...(((((((((((....)))))))...((....)))))).))))).)))))(((....))).....))))))..)......... ( -38.80) >DroSim_CAF1 1700 110 - 1 UGACUGGCUAUCCUUA----UAGGUCUAUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGGAGCCUGUGGUGAAAACCAAAUCGAUACCUGGCCCCAAAUC .....((((.......----.((((....((((((((((((((....)))))))......(.(((....))).))))))))(((....)))........))))))))....... ( -32.60) >DroEre_CAF1 1698 114 - 1 UGAAAGGGUAUCGUUAUAUGUAGGUCUAUAUAGGUUGAGUACCAAGAGGUACUCCACCAGCAUUGAGCCCAAGGAACCUGUGGUGAAAACUAAAUCAAUACCUGGCCCCAAAUC .....(((....(((.((((((....))))))(((.(((((((....))))))).)))))).....(((..(((......((((....))))........)))))))))..... ( -32.74) >DroYak_CAF1 1702 114 - 1 UGAUGGGGUUUCGUUAUAUGUAGGUCUAUAUAGGUUGAGUACCAAGAGGUACUCCACCAACAUUGAGCCCAAGGAACCUGUGGUGAAAACUAAAUCAAUACCUGGCCCCAAGUC ...(((((((..(((.((((((....))))))(((.(((((((....))))))).))))))(((((.....((....)).((((....))))..)))))....))))))).... ( -35.80) >consensus UGACUGGGUAUCCUUA____UAGGUCUAUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGGAACCUGUGGUGAAAACCAAAUCAAUACCUGGCCCCAAAUC .....((((...........(((((....((((((((((((((....)))))))......(.(((....))).))))))))(((....)))........)))))))))...... (-22.12 = -22.48 + 0.36)


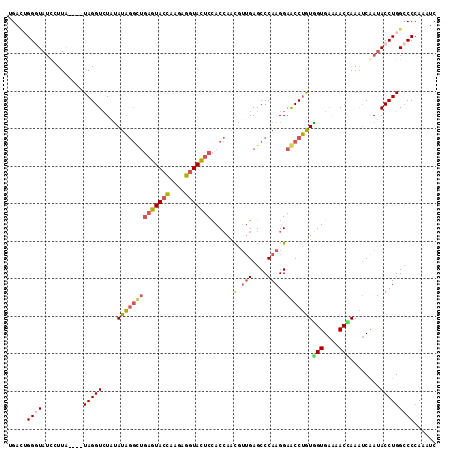
| Location | 19,933,850 – 19,933,940 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.99 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19933850 90 + 23771897 CCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAUAGACCUA----CAAGGAUACCCAAUCAGUUCC-----------AGAUUCGCUGGAGC-U-------- (((((((.((......(((.(((((((....))))))).)))......))))..----)))))..........((((((-----------((.....)))))))-)-------- ( -34.00) >DroSec_CAF1 1755 91 + 1 CCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAAAGACCUA----UAAGGAUACCCAGUCAGUUCC-----------AGAUUCGCUGGAGCCU-------- ..(((((.........(((.(((((((....))))))).)))........((..----...))...)))))...(((((-----------((.....)))))))..-------- ( -32.30) >DroSim_CAF1 1740 91 + 1 CCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAGCCUAUAUAGACCUA----UAAGGAUAGCCAGUCAGUUCC-----------AGAUUCGCUGGAGCCU-------- ....((((((......(((.(((((((....))))))).)))........((..----...)).((((.((((......-----------.)))).))))))))))-------- ( -32.50) >DroEre_CAF1 1738 95 + 1 CCUUGGGCUCAAUGCUGGUGGAGUACCUCUUGGUACUCAACCUAUAUAGACCUACAUAUAACGAUACCCUUUCAGUCUC-----------AGAUUCGCUGAAACCU-------- ...((((.((.(((..(((.(((((((....))))))).)))..))).))))))...............(((((((...-----------......)))))))...-------- ( -26.50) >DroYak_CAF1 1742 95 + 1 CCUUGGGCUCAAUGUUGGUGGAGUACCUCUUGGUACUCAACCUAUAUAGACCUACAUAUAACGAAACCCCAUCAUUCUC-----------AGAUUCGCUAAUACCU-------- ...((((.((.((((.(((.(((((((....))))))).))).)))).)))))).........................-----------................-------- ( -23.00) >DroPer_CAF1 2140 107 + 1 CCUUGGGCUCUUGGAGAGUUGAGUAUCUAUUACAGCCCAAUCGAUACACAUCUA----AAAACAAA---AAUUUGUUUUAAUUAAUUUUAACAAACUGUGAUACAUAUCUGCAU ..(((((((.....(((........))).....)))))))..((((...(((..----(((((((.---...)))))))...........((.....)))))...))))..... ( -17.50) >consensus CCUUGGGCUCAACGUUGGUGGAGUACCUCUUGGUACUCAACCUAUAUAGACCUA____UAAGGAUACCCAAUCAGUUCC___________AGAUUCGCUGAAACCU________ ...((((.((......(((.(((((((....))))))).)))......))))))............................................................ (-15.08 = -15.42 + 0.33)



| Location | 19,933,850 – 19,933,940 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.99 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19933850 90 - 23771897 --------A-GCUCCAGCGAAUCU-----------GGAACUGAUUGGGUAUCCUUG----UAGGUCUAUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGG --------(-(.(((((.....))-----------))).))..((((((.((..((----(.(((((....)))))(((((((....))))))).....)))..)))))))).. ( -30.20) >DroSec_CAF1 1755 91 - 1 --------AGGCUCCAGCGAAUCU-----------GGAACUGACUGGGUAUCCUUA----UAGGUCUUUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGG --------.(((((.((((.....-----------......((((..(((....))----).))))......((..(((((((....)))))))..))..)))))))))..... ( -30.60) >DroSim_CAF1 1740 91 - 1 --------AGGCUCCAGCGAAUCU-----------GGAACUGACUGGCUAUCCUUA----UAGGUCUAUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGG --------.(((((.((((....(-----------(((.(((...((....))...----))).))))....((..(((((((....)))))))..))..)))))))))..... ( -30.30) >DroEre_CAF1 1738 95 - 1 --------AGGUUUCAGCGAAUCU-----------GAGACUGAAAGGGUAUCGUUAUAUGUAGGUCUAUAUAGGUUGAGUACCAAGAGGUACUCCACCAGCAUUGAGCCCAAGG --------.((((((((.....))-----------))))))....((((.(((...((((((....))))))(((.(((((((....))))))).))).....))))))).... ( -33.50) >DroYak_CAF1 1742 95 - 1 --------AGGUAUUAGCGAAUCU-----------GAGAAUGAUGGGGUUUCGUUAUAUGUAGGUCUAUAUAGGUUGAGUACCAAGAGGUACUCCACCAACAUUGAGCCCAAGG --------.(((..((((((((((-----------.(......).))).)))))))((((((....))))))(((.(((((((....))))))).)))........)))..... ( -28.00) >DroPer_CAF1 2140 107 - 1 AUGCAGAUAUGUAUCACAGUUUGUUAAAAUUAAUUAAAACAAAUU---UUUGUUUU----UAGAUGUGUAUCGAUUGGGCUGUAAUAGAUACUCAACUCUCCAAGAGCCCAAGG .....((((..((((..(((((....)))))....(((((((...---.)))))))----..))))..))))..(((((((.(...(((........)))...).))))))).. ( -25.00) >consensus ________AGGCUCCAGCGAAUCU___________GAAACUGAUUGGGUAUCCUUA____UAGGUCUAUAUAGGCUGAGUACCAAGAGGUACUCCACCAACGUUGAGCCCAAGG .........(((((................................................(((((....)))))(((((((....)))))))..........)))))..... (-13.07 = -13.27 + 0.20)

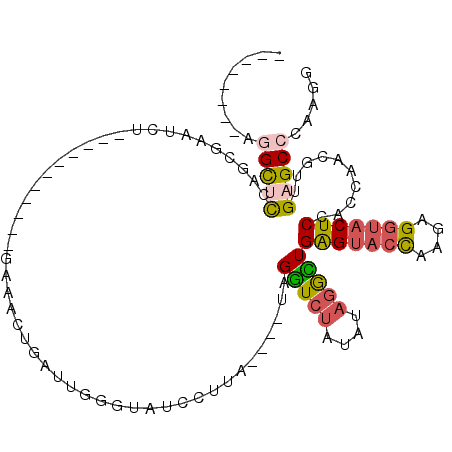

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:16 2006