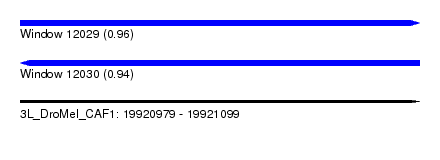
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,920,979 – 19,921,099 |
| Length | 120 |
| Max. P | 0.962208 |

| Location | 19,920,979 – 19,921,099 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -41.21 |
| Consensus MFE | -29.95 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19920979 120 + 23771897 CCGAGCAGCAUGUCCAGCUGCUCCUGCUGCACGUGCAGCAAUUGCUGUCCGUCGAGGACGAGUCAAGUCUGCCCAAUGCAGAUGGGCAAUCAGUAUCAAUGUCCAGCCAACGGAAGCAAC ..(((((((.......))))))).((((((....)))))).(((((.(((((.(.((((((......))((((((.......))))))............))))...).)))))))))). ( -47.00) >DroSec_CAF1 56717 111 + 1 CCGAGCA---------GCUGCUCCUGCUGCACGUGCAGCAAUUGCUAUCCGUCGAGGACGAGUCAAGUCUGUCCAAUGGAGAUGGGCAAUCAGUAUCAAUGUCCAGCCAACGGAAGCAAC ..((((.---------...)))).((((((....)))))).(((((.(((((...((((.((......))))))..(((...((((((...........)))))).))))))))))))). ( -42.70) >DroSim_CAF1 44054 111 + 1 CCGAGCA---------GCUGCUCCUGCUGCACGUGCAGCAAUUGCUAUCCGUCGAGGACGAGUCAAGUCUGUCCAAUGCAGAUGGGCAAUCAGUAUCAAUGUCCAGCCAACGGAAGCAAC ..((((.---------...)))).((((((....)))))).(((((.(((((.(.((((.((......))))))........((((((...........))))))..).)))))))))). ( -38.90) >DroEre_CAF1 56226 120 + 1 CCGAGCAACAUGUCCGGCUGCUCCUGCUGCACGUGCAGCAGUUGCUGUCCGUCAAGGACAACCCAAGUCUGUCCAAUGCAGAUGGGCAAUCAGUACCAAAGUCCAGCCCACGGAAGCAAC ..(((((.(......)..)))))(((((((....)))))))(((((((((.....))))...((..((((((.....))))))((((..................))))..)).))))). ( -42.47) >DroYak_CAF1 64120 120 + 1 CCGAGCAACAUGUCCAGCUGCUCUUGCUGCACGUGCAGCAGUUGCUGUCCGUCAAGGACAACUCAAGUCUGUCAAAUGCAGAUGGGUAAUCAGUAUCAAAGUCCAGCCAACGGAAGCAAC ..(((((.(.......).))))).((((((....))))))((((((.(((((...((((.......((((((.....)))))).((((.....))))...)))).....))))))))))) ( -41.30) >DroAna_CAF1 48224 105 + 1 CCCAGCAACCUUUCAAGUUGUUGCAGCUGCACCUGCAGCAAUUGCUGUCCGUCAAGGCCACCGCCAUACCGACCGAUGCCCCCGG--------------CGGCCAGCAAGUGUAA-CCAG ..(((((((.......)))))))..(((((....)))))..((((((.(((((..(((....)))....(....)........))--------------))).))))))......-.... ( -34.90) >consensus CCGAGCAACAUGUCCAGCUGCUCCUGCUGCACGUGCAGCAAUUGCUGUCCGUCAAGGACAACUCAAGUCUGUCCAAUGCAGAUGGGCAAUCAGUAUCAAUGUCCAGCCAACGGAAGCAAC ..(((((...........))))).((((((....)))))).(((((.(((((...((((..........((((((.......))))))............)))).....)))))))))). (-29.95 = -29.82 + -0.13)



| Location | 19,920,979 – 19,921,099 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -34.79 |
| Energy contribution | -35.43 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19920979 120 - 23771897 GUUGCUUCCGUUGGCUGGACAUUGAUACUGAUUGCCCAUCUGCAUUGGGCAGACUUGACUCGUCCUCGACGGACAGCAAUUGCUGCACGUGCAGCAGGAGCAGCUGGACAUGCUGCUCGG ((((((((((((((..((((............((((((.......))))))((......)))))))))))))).))))))((((((....)))))).(((((((.......))))))).. ( -55.20) >DroSec_CAF1 56717 111 - 1 GUUGCUUCCGUUGGCUGGACAUUGAUACUGAUUGCCCAUCUCCAUUGGACAGACUUGACUCGUCCUCGACGGAUAGCAAUUGCUGCACGUGCAGCAGGAGCAGC---------UGCUCGG ((((((((((((((..((((............((.(((.......))).))((......)))))))))))))).))))))((((((....)))))).((((...---------.)))).. ( -40.60) >DroSim_CAF1 44054 111 - 1 GUUGCUUCCGUUGGCUGGACAUUGAUACUGAUUGCCCAUCUGCAUUGGACAGACUUGACUCGUCCUCGACGGAUAGCAAUUGCUGCACGUGCAGCAGGAGCAGC---------UGCUCGG ((((((((((((((..((((.......(..((.((......))))..)...((......)))))))))))))).))))))((((((....)))))).((((...---------.)))).. ( -41.10) >DroEre_CAF1 56226 120 - 1 GUUGCUUCCGUGGGCUGGACUUUGGUACUGAUUGCCCAUCUGCAUUGGACAGACUUGGGUUGUCCUUGACGGACAGCAACUGCUGCACGUGCAGCAGGAGCAGCCGGACAUGUUGCUCGG .((((((((((((((.((((....)).))....)))).....((..((((((.(....))))))).))))))).)))))(((((((....)))))))(((((((.......))))))).. ( -49.40) >DroYak_CAF1 64120 120 - 1 GUUGCUUCCGUUGGCUGGACUUUGAUACUGAUUACCCAUCUGCAUUUGACAGACUUGAGUUGUCCUUGACGGACAGCAACUGCUGCACGUGCAGCAAGAGCAGCUGGACAUGUUGCUCGG (((((((((((..(..((((..((((....))))....(((((....).))))........)))))..))))).))))))((((((....)))))).(((((((.......))))))).. ( -45.50) >DroAna_CAF1 48224 105 - 1 CUGG-UUACACUUGCUGGCCG--------------CCGGGGGCAUCGGUCGGUAUGGCGGUGGCCUUGACGGACAGCAAUUGCUGCAGGUGCAGCUGCAACAACUUGAAAGGUUGCUGGG (..(-(.((..((((((.(((--------------.(((((.(((((.((.....))))))).))))).))).))))))..(((((....)))))................)).))..). ( -42.20) >consensus GUUGCUUCCGUUGGCUGGACAUUGAUACUGAUUGCCCAUCUGCAUUGGACAGACUUGACUCGUCCUCGACGGACAGCAAUUGCUGCACGUGCAGCAGGAGCAGCUGGACAUGUUGCUCGG ((((((((((((((..((((...............(((.......)))...((......)))))))))))))).))))))((((((....)))))).(((((((.......))))))).. (-34.79 = -35.43 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:11 2006