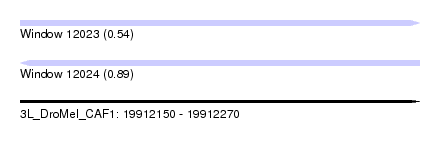
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,912,150 – 19,912,270 |
| Length | 120 |
| Max. P | 0.894492 |

| Location | 19,912,150 – 19,912,270 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -28.52 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19912150 120 + 23771897 UCGAAAGGAGCUUGCUCAGCAUUCUCGGGAUCUCGAGUAGCCCCGCGGCCAUGGCUACACUCGUAUGUAUACACACGACGCAUGCGCAGACCUCGGGACUCCGCCGAGAUCCCUCCUCCA .(....)(((..(((...)))..)))(((((((((.(.((.((((.((...((((.....((((.((....)).)))).....)).))..)).)))).)).)..)))))))))....... ( -43.70) >DroSec_CAF1 48011 120 + 1 UCGAAAGGAGCUGGCUCAGCAUUCUCGGGAUCGCGAGUAGCCCCAUGGCCAUGGCUGCACUCGUAUGUAUACACACGACGCAUGCGGAGACCUCGGGACUCCGCCGAGAUCCCUACUCCA ......(((((((...))))......(((((((((.((((((..........))))))..((((.((....)).)))))))..((((((.((...)).))))))...))))))...))). ( -46.80) >DroSim_CAF1 32376 120 + 1 UCGAAAGGAGCUGGCUCAGCAUUCUCGGGAUCGCGAGUAGCCCCAUGGACAUGGCUACACUCGUAUGUAUACACACGACGCAUGCGCAGACCUCGGGACUCCGCCGAGAUCCCUACUCCA ......((((..(((((.((..(((((((.(((((.((((((..........))))))..((((.((....)).))))......))).)).)))))))....)).)))...))..)))). ( -40.00) >DroEre_CAF1 47224 106 + 1 UCGAAAGGAGCUGGCUCAGCGUUCUCGGGAUCUCGAGUAGC--------CGUGGCUACA------AGUAUACACACGACGCAUGCGCAGAGCUCGAGACUCCGCCGAGAUCGCUCCUCCA .....((((((...(((.(((.(((((((.((((..(((((--------....))))).------.((((.(.......).))))).))).)))))))...))).)))...))))))... ( -40.30) >DroYak_CAF1 47133 106 + 1 UCGAAAGGAGCUGGCUCAGCAUUCUCGGGAUCUCGAGUAGC--------CAUGGCUACA------UGUAUACACUCAACGCAUGCGCAGAUCGCGAGACUCCGAUGAGAUCCCUCCUCCA ......((((..((((((....(((((.(((((((.(((((--------....)))))(------(((...........)))).)).))))).)))))......))))...))..)))). ( -37.10) >consensus UCGAAAGGAGCUGGCUCAGCAUUCUCGGGAUCUCGAGUAGCCCC__GG_CAUGGCUACACUCGUAUGUAUACACACGACGCAUGCGCAGACCUCGGGACUCCGCCGAGAUCCCUCCUCCA ......(((((((...))))......(((((((((.((((((..........)))))).........................(((.((.(.....).)).))))))))))))...))). (-28.52 = -29.68 + 1.16)

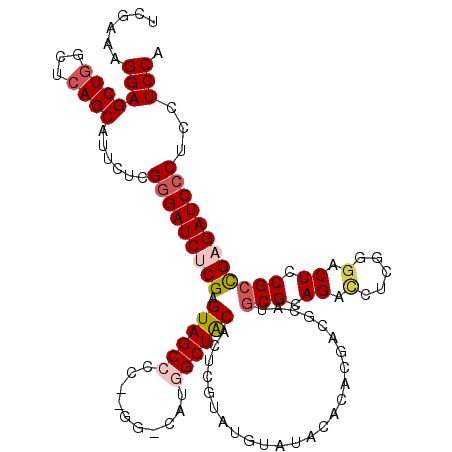
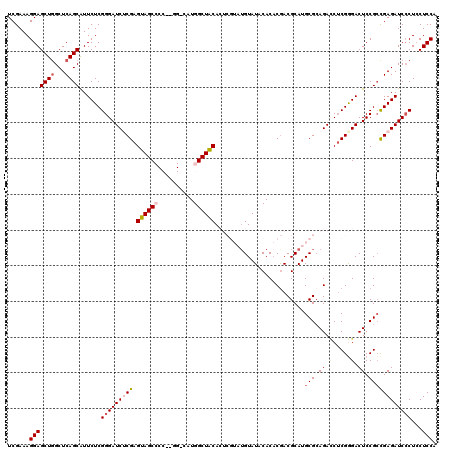
| Location | 19,912,150 – 19,912,270 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -52.28 |
| Consensus MFE | -33.24 |
| Energy contribution | -39.00 |
| Covariance contribution | 5.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19912150 120 - 23771897 UGGAGGAGGGAUCUCGGCGGAGUCCCGAGGUCUGCGCAUGCGUCGUGUGUAUACAUACGAGUGUAGCCAUGGCCGCGGGGCUACUCGAGAUCCCGAGAAUGCUGAGCAAGCUCCUUUCGA .(((((.((((((((((...(((((((.((((((.((.(((((((((((....))))))).)))))))).)))).)))))))..))))))))))(((..(((...)))..)))))))).. ( -63.30) >DroSec_CAF1 48011 120 - 1 UGGAGUAGGGAUCUCGGCGGAGUCCCGAGGUCUCCGCAUGCGUCGUGUGUAUACAUACGAGUGCAGCCAUGGCCAUGGGGCUACUCGCGAUCCCGAGAAUGCUGAGCCAGCUCCUUUCGA .(((((.((...(((((((.(((((((.((((...((.(((((((((((....))))))).))))))...)))).))))))).((((......))))..))))))))).)))))...... ( -56.70) >DroSim_CAF1 32376 120 - 1 UGGAGUAGGGAUCUCGGCGGAGUCCCGAGGUCUGCGCAUGCGUCGUGUGUAUACAUACGAGUGUAGCCAUGUCCAUGGGGCUACUCGCGAUCCCGAGAAUGCUGAGCCAGCUCCUUUCGA .(((((.((...(((((((.(((((((.((.(((.((.(((((((((((....))))))).)))))))).).)).))))))).((((......))))..))))))))).)))))...... ( -52.60) >DroEre_CAF1 47224 106 - 1 UGGAGGAGCGAUCUCGGCGGAGUCUCGAGCUCUGCGCAUGCGUCGUGUGUAUACU------UGUAGCCACG--------GCUACUCGAGAUCCCGAGAACGCUGAGCCAGCUCCUUUCGA .((((((((...(((((((...(((((..(((((((((((...))))))))....------.(((((....--------)))))..)))....))))).)))))))...))))))))... ( -49.70) >DroYak_CAF1 47133 106 - 1 UGGAGGAGGGAUCUCAUCGGAGUCUCGCGAUCUGCGCAUGCGUUGAGUGUAUACA------UGUAGCCAUG--------GCUACUCGAGAUCCCGAGAAUGCUGAGCCAGCUCCUUUCGA ....(((((((.((..((((..(((((.(((((.((.(((((.....)))))...------.(((((....--------))))).))))))).)))))...))))...)).))))))).. ( -39.10) >consensus UGGAGGAGGGAUCUCGGCGGAGUCCCGAGGUCUGCGCAUGCGUCGUGUGUAUACAUACGAGUGUAGCCAUG_CC__GGGGCUACUCGAGAUCCCGAGAAUGCUGAGCCAGCUCCUUUCGA .(((...((((((((((...(((((((.((.(((.((.(((((((((((....))))))).)))))))).).)).)))))))..))))))))))......((((...)))))))...... (-33.24 = -39.00 + 5.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:05 2006