| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,905,133 – 19,905,225 |
| Length | 92 |
| Max. P | 0.630330 |

| Location | 19,905,133 – 19,905,225 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.24 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
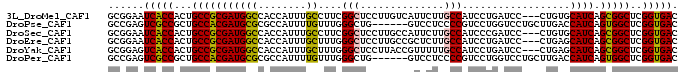
>3L_DroMel_CAF1 19905133 92 + 23771897 GUCACCGAGCCGCUGAUGCACAG---GGAUCAGGAUGGCAAGAAUGACAAGGAGCCGAAGGCAAAUGGUGGCCAUCGCGGCAGUGGUGAUUCCGC (((((((.(((((((((.(....---).)))).((((((..(.....).....(((...)))........)))))))))))..)))))))..... ( -33.60) >DroPse_CAF1 35458 89 + 1 GUCACCGAGCCACUGAUGGUCAAGCAGGACCAGGACGGGGAGGAC------CAGCCCAAACAAAAUGGCGCGCAUCGUGGCAGCGGCGACUCGGC (((.(((.(((((.(((((((...(....((.....)).)..)))------).(((((.......))).))..))))))))..))).)))..... ( -31.00) >DroSec_CAF1 41354 92 + 1 GUCACCGAGCCGCUGAUGCACAG---GGAUCGGGAUGGCAAGAAUGGCAAGGAGCCGAAGGCAAAUGGUGGCCAUCGCGGCAGUGGUGAUUCCGC (((((((.(((((((((.(....---).)))).((((((.....((((.....))))...((.....)).)))))))))))..)))))))..... ( -36.10) >DroEre_CAF1 40235 92 + 1 GUCACCGAGCCGCUGAUGCUCAG---GGAUCAGGAUGGCAAGAGCGGCAAGGAGCCCAAAGCAAAUGGUGGCCAUCGCGGCAGUGGUGAUUCCGC (((((((.(((((((((.(....---).)))).((((((....(.(((.....))))...((.....)).)))))))))))..)))))))..... ( -36.50) >DroYak_CAF1 40418 92 + 1 GUCACCGAGCCGCUGAUGCUCAG---GGAUCAGGAUGGCAAAAACGGUAAGGAGCCCAAAGCAAAUGGUGGCCAUCGCGGCAGUGGUGACUCCGC (((((((.(((((((((.(....---).)))).((((((......(((.....)))....((.....)).)))))))))))..)))))))..... ( -36.70) >DroPer_CAF1 35549 89 + 1 GUCACCGAGCCACUGAUGGUCAAGCAGGACCAGGACGGGGAGGAC------CAGCCCAAACAAAAUGGCGCGCAUCGUGGCAGCGGCGACUCGGC (((.(((.(((((.(((((((...(....((.....)).)..)))------).(((((.......))).))..))))))))..))).)))..... ( -31.00) >consensus GUCACCGAGCCGCUGAUGCUCAG___GGAUCAGGAUGGCAAGAACGGCAAGGAGCCCAAAGCAAAUGGUGGCCAUCGCGGCAGUGGUGACUCCGC (((((((.(((((((((.(.......).)))).((((((..............(((..........))).)))))))))))..)))))))..... (-25.30 = -25.24 + -0.05)

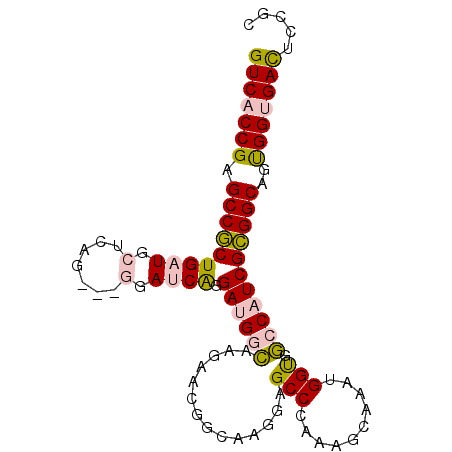
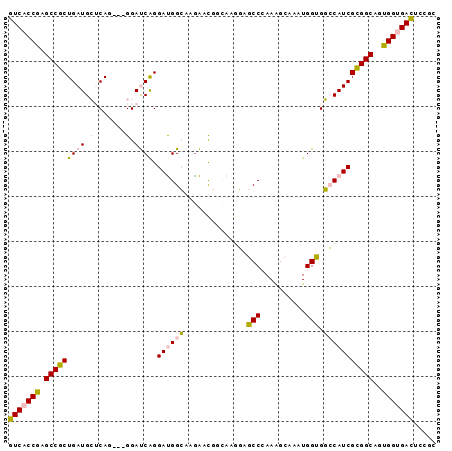
| Location | 19,905,133 – 19,905,225 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -23.08 |
| Energy contribution | -21.86 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19905133 92 - 23771897 GCGGAAUCACCACUGCCGCGAUGGCCACCAUUUGCCUUCGGCUCCUUGUCAUUCUUGCCAUCCUGAUCC---CUGUGCAUCAGCGGCUCGGUGAC ......(((((...(((((((((((...((...(((...)))....))........)))))).((((..---......)))))))))..))))). ( -28.40) >DroPse_CAF1 35458 89 - 1 GCCGAGUCGCCGCUGCCACGAUGCGCGCCAUUUUGUUUGGGCUG------GUCCUCCCCGUCCUGGUCCUGCUUGACCAUCAGUGGCUCGGUGAC .....(((((((..(((((((((.((.(((.......))))).(------(....)).)))).(((((......)))))...))))).))))))) ( -34.20) >DroSec_CAF1 41354 92 - 1 GCGGAAUCACCACUGCCGCGAUGGCCACCAUUUGCCUUCGGCUCCUUGCCAUUCUUGCCAUCCCGAUCC---CUGUGCAUCAGCGGCUCGGUGAC ......(((((...(((((((((((((.....)).....(((.....)))......))))))..(((..---......))).)))))..))))). ( -30.60) >DroEre_CAF1 40235 92 - 1 GCGGAAUCACCACUGCCGCGAUGGCCACCAUUUGCUUUGGGCUCCUUGCCGCUCUUGCCAUCCUGAUCC---CUGAGCAUCAGCGGCUCGGUGAC ......(((((...((((((((((...)))))(((((.(((.((......((....))......)).))---).)))))...)))))..))))). ( -33.60) >DroYak_CAF1 40418 92 - 1 GCGGAGUCACCACUGCCGCGAUGGCCACCAUUUGCUUUGGGCUCCUUACCGUUUUUGCCAUCCUGAUCC---CUGAGCAUCAGCGGCUCGGUGAC .....((((((...((((((((((...)))))(((((.(((.((......((....))......)).))---).)))))...)))))..)))))) ( -33.60) >DroPer_CAF1 35549 89 - 1 GCCGAGUCGCCGCUGCCACGAUGCGCGCCAUUUUGUUUGGGCUG------GUCCUCCCCGUCCUGGUCCUGCUUGACCAUCAGUGGCUCGGUGAC .....(((((((..(((((((((.((.(((.......))))).(------(....)).)))).(((((......)))))...))))).))))))) ( -34.20) >consensus GCGGAAUCACCACUGCCGCGAUGGCCACCAUUUGCUUUGGGCUCCUUGCCGUUCUUGCCAUCCUGAUCC___CUGAGCAUCAGCGGCUCGGUGAC ......(((((...(((((((((((........))....(((..............)))..................)))).)))))..))))). (-23.08 = -21.86 + -1.22)
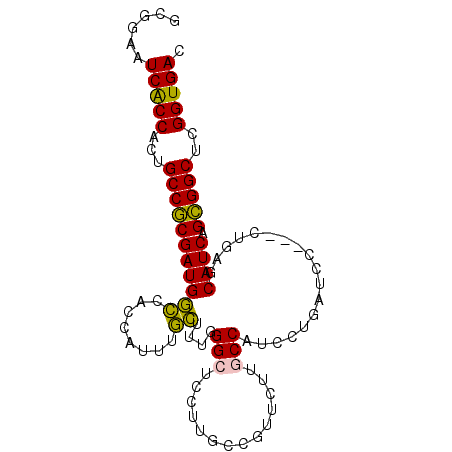
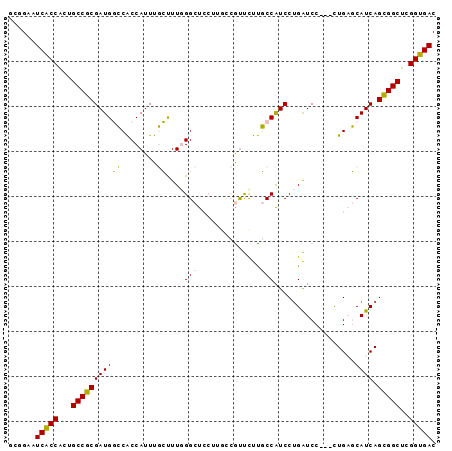
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:00 2006