
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,882,102 – 19,882,195 |
| Length | 93 |
| Max. P | 0.734460 |

| Location | 19,882,102 – 19,882,195 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.56 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -10.81 |
| Energy contribution | -11.53 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
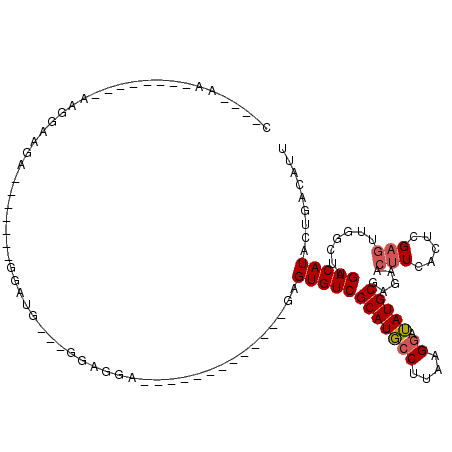
>3L_DroMel_CAF1 19882102 93 + 23771897 U----AAAAGAAAAAAAGGACGA-------GGAUG---GAAGGA-------------GAGUGUCGCAUGCCUUAAGGAUAUGCGAGAACUUCAGUCGAGGCGGCUUGACAUACUGACAUU .----..................-------.((((---..((((-------------(..(.(((((((((....)).))))))).)..))).((((((....))))))...))..)))) ( -25.90) >DroPse_CAF1 15849 98 + 1 G----GA--------GGGGGAACUAUGUAUGUGUGCGCGGAGGA---GUG-------CCGUGUCGCAUGCCUGCAGGAUAUGCGACAACUUUGCUUGAGUUGCCCCGACAUACUGACAUU (----..--------((((.((((..(((.(((.((((......---)))-------)).(((((((((((....)).)))))))))))..)))...)))).))))..)........... ( -38.50) >DroSec_CAF1 17678 92 + 1 U----AAAAGGAA-AAAGGACGA-------GGAUG---GAAGGA-------------GAGUGUCGCAUGCCUUAAGUAUAUGCGAGAACUUCAGUCGAGCCGGCUUGACAUACUGACAUU .----........-.........-------.((((---..((((-------------(..(.(((((((.........))))))).)..))).((((((....))))))...))..)))) ( -22.50) >DroEre_CAF1 16691 79 + 1 C----GA--------AAGACAGA-------GGAUG----GAGCA-------------GAGUGUCGCAUGCCUUCGGGAUAUGCGAGAACUUCAAGCG-----ACUUGACAUACUGACAUU (----..--------..).....-------.((((----...((-------------(.((((((((((((....)).))))))).....(((((..-----.)))))))).))).)))) ( -23.80) >DroAna_CAF1 14158 107 + 1 CGGUUAG-----A-AGAGGCAGUGGU----GUAUG---GGAGGAGGGGUUCCUGAAGGAGUGUCGCAUUCCUUAAGGAAAUGCGAGAACUUCACUCGAGUUGGAUAGACAUACUGAUGUU .......-----.-.....(((((((----.((((---((((......)))))...(((((.(((((((((....)).)))))))..)))))...........))).)).)))))..... ( -28.70) >DroPer_CAF1 15518 94 + 1 G----GA--------GGGGGAACUAU----GUGUGCGCGGAGGA---GUG-------CCGUGUCGCAUGCCUGCAGGAUAUGCGACAACUUUGCUUGAGUUGUCCCGACAUACUGACAUU (----..--------((((.((((..----(((.((((......---)))-------)).(((((((((((....)).))))))))).....))...)))).))))..)........... ( -34.80) >consensus C____AA________AAGGAAGA_______GGAUG___GGAGGA_____________GAGUGUCGCAUGCCUUAAGGAUAUGCGAGAACUUCACUCGAGUUGGCUUGACAUACUGACAUU ...........................................................((((((((((((....)).))))).....(((.....))).......)))))......... (-10.81 = -11.53 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:58 2006