
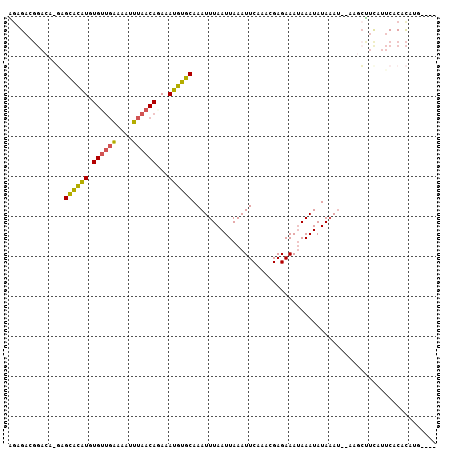
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,875,886 – 19,876,001 |
| Length | 115 |
| Max. P | 0.820707 |

| Location | 19,875,886 – 19,875,987 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -14.67 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.37 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19875886 101 - 23771897 AGCGACGGACAUGAGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUAAAU--AAGCUUUAUUCACACAUG---- .((...........)).(((((((.(((......(((....)))((..((((((((.(.(((......))).).))).)))))--..))....))))))))))---- ( -15.00) >DroSec_CAF1 11664 101 - 1 AGCGACGGACAUGGGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUAAAU--AAGCUUCAUUCACACAUG---- .((..((....)).)).(((((((.(((......(((....)))((..((((((((.(.(((......))).).))).)))))--..))....))))))))))---- ( -15.30) >DroEre_CAF1 10945 100 - 1 GCAGACGGACA-GAGCACAUGUGUUGAAAAUUUAACAUAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUAAAU--AAGCCUCAUUCACACAUG---- ...((.((.(.-..(((((((((((((....)))))))..)))))).............(((......)))............--..)))))...........---- ( -15.40) >DroWil_CAF1 16189 76 - 1 -----------UGUGUGUGUGUGUCUGAAAUUUAACACAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAGAAAUAUAUAC--AUAU------------------ -----------(((((((((((.(((.((((((..(((....))).)))))).......(((......)))))).))))))))--))).------------------ ( -15.40) >DroYak_CAF1 11099 104 - 1 GGAGACGGACA-GAGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUAAAU--AAGCUUUAUUCACACAUUUAUU (....)(((.(-((((...((((((..((((((.(((....)))..)))))).......(((......)))...))))))...--..))))).)))........... ( -14.50) >DroAna_CAF1 8823 91 - 1 AGAG----------GCACAUGUGGUGAAAAUUUAACACAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUAAAUAUACGCUUCGUUCGCAUA------ ....----------((((((...(((.........)))..)))))).................((((((....................))))))......------ ( -12.45) >consensus AGAGACGGACA_GAGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUAAAU__AAGCUUCAUUCACACAUG____ ..............((((((.((((((....))))))...))))))............................................................. ( -9.56 = -9.37 + -0.19)


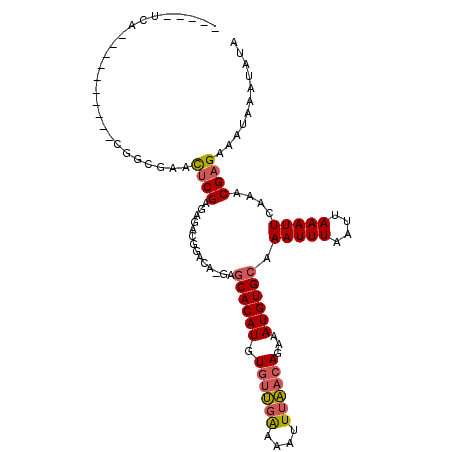
| Location | 19,875,907 – 19,876,001 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -16.24 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19875907 94 - 23771897 -----UUG----------CGGCGAACUCGAGCGACGGACAUGAGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA -----(((----------(..((....)).)))).........((((((.((((((....))))))...)))))).................................. ( -16.60) >DroVir_CAF1 13152 107 - 1 AUCCGUCACAGACACG--CCCCGUGUUCGUCACAUAGAAAGCGACACAUGUAUAUGAAAUUUUACACAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA .(((((....((((((--...)))))).(((.(.......).)))...(((((((..............))))))).................))).)).......... ( -13.24) >DroSec_CAF1 11685 94 - 1 -----UUG----------CGGCGAACUCGAGCGACGGACAUGGGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA -----...----------.......((((..((((..(((((....))))))))).((((((.(((....)))..)))))).............))))........... ( -17.60) >DroEre_CAF1 10966 93 - 1 -----CCA----------CGGCAAACGCGGCAGACGGACA-GAGCACAUGUGUUGAAAAUUUAACAUAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA -----((.----------((.....)).))..........-..(((((((((((((....)))))))..)))))).................................. ( -15.90) >DroYak_CAF1 11124 100 - 1 -----UCACAA---AACUCGGCAAACUCGGGAGACGGACA-GAGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA -----......---..((((......((.(....).))..-..((((((.((((((....))))))...))))))...................))))........... ( -21.40) >DroAna_CAF1 8844 83 - 1 -----UCG----------CA-AGUAUUCGAGAG----------GCACAUGUGGUGAAAAUUUAACACAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA -----.(.----------..-.)..((((....----------((((((...(((.........)))..)))))).(((((....)))))....))))........... ( -12.70) >consensus _____UCA__________CGGCGAACUCGAGAGACGGACA_GAGCACAUGUGUUGAAAAUUUAACAGAAAUGUGCAAAUUUAAUUAAAUUCAAACGAGAAAUAAAUAUA .........................((((..............((((((.((((((....))))))...)))))).(((((....)))))....))))........... (-11.70 = -11.90 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:57 2006