
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,227,724 – 2,227,830 |
| Length | 106 |
| Max. P | 0.999807 |

| Location | 2,227,724 – 2,227,830 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.16 |
| Mean single sequence MFE | -37.56 |
| Consensus MFE | -26.39 |
| Energy contribution | -25.71 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
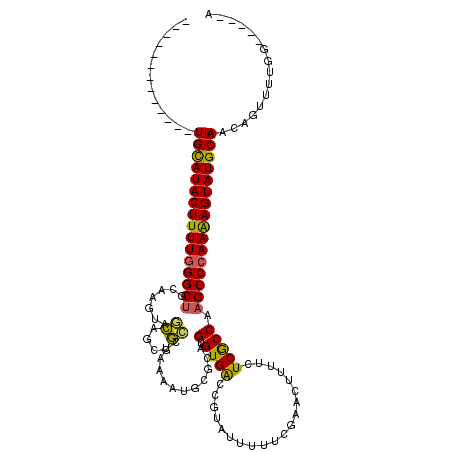
>3L_DroMel_CAF1 2227724 106 + 23771897 -------------UGCAUACUUUUGGGCUGCACGUAUCUGCGGAGUGAACUGUGCAGUGUGGCCGUAUUUUUUGAACUGUUCUCACCAAGCCCAAAAGUAUGCAACAGUUUUGGGUGA-A -------------((((((((((((((((.((((...(((((.((....)).)))))))))...........(((.......)))...))))))))))))))))..............-. ( -39.50) >DroSec_CAF1 8701 98 + 1 -------------UGCAUACUUUUGGGCUGCAAGUAGCCGCGGCCUUAAGUGAGCAGUGUGACCGUAUUUUUCGAAUUUUUCUCGCCAAGCCCAAAAGUAUGCAACAGUUU--------- -------------((((((((((((((((.......(((((........))).)).(((.((....((((...))))...)).)))..)))))))))))))))).......--------- ( -34.00) >DroSim_CAF1 8809 98 + 1 -------------UGCAUACUUUUGGGCUGCAAGUAGCCGCGGCCUAAAGUGCGCAGUGUGACCGUAUUAUUCGAAUUUUUCUCACCAAGCCCAAAAGUAUGCAACAGUUU--------- -------------(((((((((((((((((((..((((....).)))...)))...(((.((....(((.....)))...)).)))..)))))))))))))))).......--------- ( -35.00) >DroEre_CAF1 21634 118 + 1 CGGCAAAUAAUGUUGUAUACUUUUGGGCUGCAAAUGGUAACGAGCUAAAAUGUCCAGUGUGACCAUAUUUUCCGA-CUUAUGUCGCCAAGCCCAAAAGUAUGCAACAGGGUUGGCGGC-A ((.(((....(((((((((((((((((((((..(((((.(((..(......)..).))...))))).......((-(....)))))..)))))))))))))))))))...))).))..-. ( -48.50) >DroYak_CAF1 8916 116 + 1 ----AAAUCGUGUUGCAUACUUUUAGGCACCAAAUUGCAACGGCCUAGAAUACAUAGUGUGACCAUGUUUUUCGAACUGUUCUCGCCAAGCCCAAGAGUAUGCAACAAUUUUGGGGCAAA ----......((((((((((((((.(((..(.....)....(((..(((....(((((.(((.........))).)))))))).)))..))).))))))))))))))............. ( -30.80) >consensus _____________UGCAUACUUUUGGGCUGCAAGUAGCAGCGGCCUAAAAUGCGCAGUGUGACCGUAUUUUUCGAACUUUUCUCGCCAAGCCCAAAAGUAUGCAACAGUUUUGG_____A .............((((((((((((((((.......((....))............(.((((....................))))).))))))))))))))))................ (-26.39 = -25.71 + -0.68)

| Location | 2,227,724 – 2,227,830 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.16 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
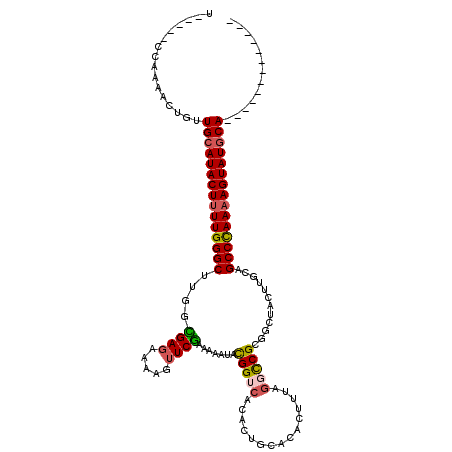
>3L_DroMel_CAF1 2227724 106 - 23771897 U-UCACCCAAAACUGUUGCAUACUUUUGGGCUUGGUGAGAACAGUUCAAAAAAUACGGCCACACUGCACAGUUCACUCCGCAGAUACGUGCAGCCCAAAAGUAUGCA------------- .-..............((((((((((((((((.((.(.((((.((.........)).((......))...)))).).))(((......)))))))))))))))))))------------- ( -35.20) >DroSec_CAF1 8701 98 - 1 ---------AAACUGUUGCAUACUUUUGGGCUUGGCGAGAAAAAUUCGAAAAAUACGGUCACACUGCUCACUUAAGGCCGCGGCUACUUGCAGCCCAAAAGUAUGCA------------- ---------.......((((((((((((((((..(((((................(((.....))).........(((....))).)))))))))))))))))))))------------- ( -36.80) >DroSim_CAF1 8809 98 - 1 ---------AAACUGUUGCAUACUUUUGGGCUUGGUGAGAAAAAUUCGAAUAAUACGGUCACACUGCGCACUUUAGGCCGCGGCUACUUGCAGCCCAAAAGUAUGCA------------- ---------.......((((((((((((((((..(..((......(((.......))).....(((((..(....)..)))))...))..)))))))))))))))))------------- ( -36.10) >DroEre_CAF1 21634 118 - 1 U-GCCGCCAACCCUGUUGCAUACUUUUGGGCUUGGCGACAUAAG-UCGGAAAAUAUGGUCACACUGGACAUUUUAGCUCGUUACCAUUUGCAGCCCAAAAGUAUACAACAUUAUUUGCCG .-..((.(((...(((((.(((((((((((((..(((((....)-)).......(((((.((.(((((...)))))...)).)))))..))))))))))))))).)))))....))).)) ( -35.70) >DroYak_CAF1 8916 116 - 1 UUUGCCCCAAAAUUGUUGCAUACUCUUGGGCUUGGCGAGAACAGUUCGAAAAACAUGGUCACACUAUGUAUUCUAGGCCGUUGCAAUUUGGUGCCUAAAAGUAUGCAACACGAUUU---- .............(((((((((((.((((((((((((((.....))))....((((((.....))))))...))))((((........)))))))))).)))))))))))......---- ( -35.30) >consensus U_____CCAAAACUGUUGCAUACUUUUGGGCUUGGCGAGAAAAGUUCGAAAAAUACGGUCACACUGCACACUUUAGGCCGCGGCUACUUGCAGCCCAAAAGUAUGCA_____________ ................(((((((((((((((....((((.....)))).......(((((...............)))))............)))))))))))))))............. (-26.50 = -26.74 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:51 2006