| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,844,693 – 19,844,804 |
| Length | 111 |
| Max. P | 0.976328 |

| Location | 19,844,693 – 19,844,804 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -26.00 |
| Energy contribution | -25.92 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
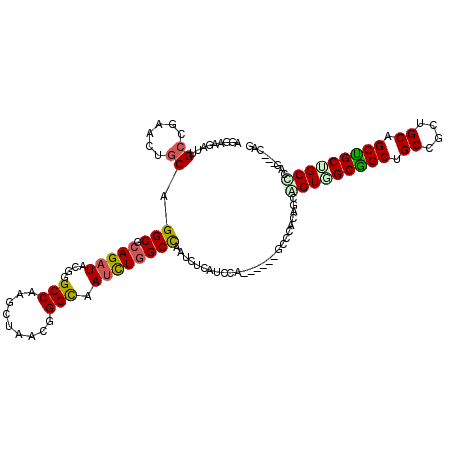
>3L_DroMel_CAF1 19844693 111 + 23771897 AGGAAGAUUGCCGAGUUGCAGGCACAGAUCAGGGCCAAGCUUACUGGCAAUUUGGCUAGUUUGAUUCA------GCCCACUGCGGUAGCGGCCGCCGCUGCAGCUGCUGCUCAA---GCG .........((((((((((.(((.(......).))).((....)).))))))))))..((((((..((------((...((((((..((....))..))))))..)))).))))---)). ( -44.00) >DroPse_CAF1 30241 111 + 1 AGAAAGAUCGCAGAGCUGCAGGCCCAGAUACGUGCCAAGCUAACGGGCAAUCUGGCCAACCUCAUCCA------ACCCACAGCCGUGGCGGCUGCCGCUGCAGCUGCUGCCCAG---CAG .........(((((((((((((((.((((...((((.........))))))))))))...........------.....((((((...))))))....))))))).))))....---... ( -42.20) >DroGri_CAF1 31734 120 + 1 AAGAAGAUUGCCGAAUUGCAGGCGCAGAUACGCGCCAAGCUAACGGGCAAUUUGGCCAAUCUUAUACAGCAGCAGCCAACGGCGGUUGCAGCUGCUGCUGCCGCUGCCGCCCAGCAACAG ...((((((((((((((((.(((((......)))))...(....).))))))))).)))))))...((((((((((((((....))))..))))))))))..((((.....))))..... ( -57.80) >DroYak_CAF1 28548 111 + 1 AGGAAGAUUGCCGAAUUGCAGGCGCAGAUCAGGGCCAAGCUUACUGGUAAUUUGGCCAGUUUGAUUCA------GCCAACUGCAGUGGCGGCCGCCGCUGCAGCCGCCGCUCAG---GCG .((.....(((......)))(((...(((((.(((((((.(((....))))))))))....)))))..------)))..((((((((((....))))))))))))(((.....)---)). ( -47.90) >DroAna_CAF1 28985 108 + 1 AGGAAGAUAGCCGAACUACAGGCGCAGAUUCGGGCCAAACUAACUGGCAACCUAGCUAGUCUCAUCCA------GCCCU---CAGUAGCGGCAGCAGCUGCCGCUGCGGCUCAA---GCU ..(((....(((........))).....)))(((((......((((((......))))))........------.....---..((((((((((...)))))))))))))))..---... ( -41.90) >DroPer_CAF1 30479 111 + 1 AGAAAGAUCGCAGAGCUGCAGGCCCAGAUACGUGCCAAGCUAACGGGCAAUCUGGCCAACCUCAUCCA------ACCCACAGCCGUGGCGGCUGCCGCUGCAGCUGCUGCCCAG---CAG .........(((((((((((((((.((((...((((.........))))))))))))...........------.....((((((...))))))....))))))).))))....---... ( -42.20) >consensus AGGAAGAUUGCCGAACUGCAGGCGCAGAUACGGGCCAAGCUAACGGGCAAUCUGGCCAAUCUCAUCCA______GCCCACAGCAGUGGCGGCUGCCGCUGCAGCUGCUGCCCAG___CAG .........((......)).(((.(((((....(((.........))).))))))))..........................(((((((((.((....)).)))))))))......... (-26.00 = -25.92 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:43 2006