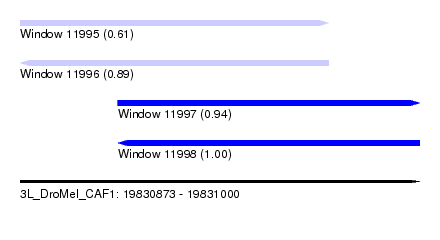
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,830,873 – 19,831,000 |
| Length | 127 |
| Max. P | 0.999956 |

| Location | 19,830,873 – 19,830,971 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -11.81 |
| Energy contribution | -13.65 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19830873 98 + 23771897 CCGCUCUCUUUAUCGCUGCAGCGAUAAAGUUCCAACAA----AACAGAAAGAAA--AACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUU .......(((((((((....))))))))).........----............--((((((.((((((((...((....)).)).))))))......)))))) ( -26.20) >DroVir_CAF1 15304 89 + 1 UUUCUCGGUCGCUCGGCACGCAGAAAACCUUUCACUGCCCCGCGCACACCCAAAAAAACA---------------CAACAGCCAACCAGGCACAAUCAGCUGUG .....((((.((.(((...((((...........)))).))).))...............---------------.....(((.....))).......)))).. ( -17.00) >DroSec_CAF1 14551 98 + 1 CCGCUCUCUUUAUCGCUGCAGCGAAAAAGUUCCAACAA----AACAGAAAGAAA--AACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUU .......((((.((((....)))).)))).........----............--((((((.((((((((...((....)).)).))))))......)))))) ( -22.10) >DroSim_CAF1 17794 98 + 1 CCGCUCUCUUUAUCGCUGCAGCGAAAAAGUUCCAACAA----AACAGAAAGAAA--AACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUU .......((((.((((....)))).)))).........----............--((((((.((((((((...((....)).)).))))))......)))))) ( -22.10) >DroEre_CAF1 14511 95 + 1 CCGCUCUCUUUAUCGCUGCAGCGAAAAAGUUUCAACAA----AACAGAAAGAAA--AACAGCACGACUGCAA---CAACCGUGUGCCAGUCGCAAUCAGCUGUU ......(((((.((((....))))....((((.....)----)))..)))))..--((((((.((((((..(---((....)))..))))))......)))))) ( -22.30) >DroYak_CAF1 14423 95 + 1 CCGCUCUCUUUAUCGCUGCAGCGAAGAAGUUCCAACAA----AACAGAAAUAAA--AACAGCACGACUGCAA---CAACCGUGUGCCAGUCGCAAUCAGCUGUU .......((((.((((....)))).)))).........----............--((((((.((((((..(---((....)))..))))))......)))))) ( -21.80) >consensus CCGCUCUCUUUAUCGCUGCAGCGAAAAAGUUCCAACAA____AACAGAAAGAAA__AACAGCACGACUGCAA___CAACCGUGUGCCAGUCGCAAUCAGCUGUU .......((((.((((....)))).))))...........................((((((.((((((((............)).))))))......)))))) (-11.81 = -13.65 + 1.84)

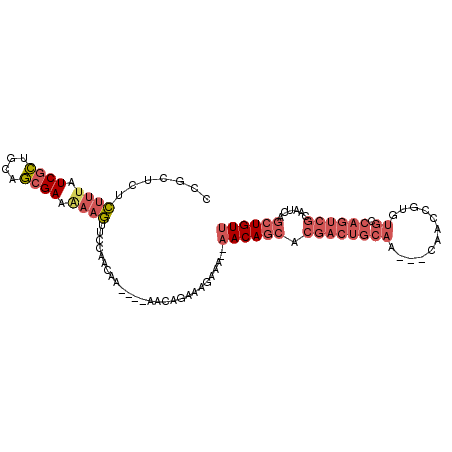

| Location | 19,830,873 – 19,830,971 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -18.46 |
| Energy contribution | -20.43 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19830873 98 - 23771897 AACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUU--UUUCUUUCUGUU----UUGUUGGAACUUUAUCGCUGCAGCGAUAAAGAGAGCGG ((((((((((((((((.((((......)))))))))))))).))))))--.......(((((----((.......(((((((((....)))))))))))))))) ( -36.41) >DroVir_CAF1 15304 89 - 1 CACAGCUGAUUGUGCCUGGUUGGCUGUUG---------------UGUUUUUUUGGGUGUGCGCGGGGCAGUGAAAGGUUUUCUGCGUGCCGAGCGACCGAGAAA .(((((..(((......)))..)))))..---------------...((((((((...(((.(((.((.(((((.....))).)))).))).))).)))))))) ( -29.40) >DroSec_CAF1 14551 98 - 1 AACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUU--UUUCUUUCUGUU----UUGUUGGAACUUUUUCGCUGCAGCGAUAAAGAGAGCGG ((((((((((((((((.((((......)))))))))))))).))))))--...((((((...----((((((.(.(......).).))))))...))))))... ( -33.50) >DroSim_CAF1 17794 98 - 1 AACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUU--UUUCUUUCUGUU----UUGUUGGAACUUUUUCGCUGCAGCGAUAAAGAGAGCGG ((((((((((((((((.((((......)))))))))))))).))))))--...((((((...----((((((.(.(......).).))))))...))))))... ( -33.50) >DroEre_CAF1 14511 95 - 1 AACAGCUGAUUGCGACUGGCACACGGUUG---UUGCAGUCGUGCUGUU--UUUCUUUCUGUU----UUGUUGAAACUUUUUCGCUGCAGCGAUAAAGAGAGCGG ((((((((((((((((.(((.....))))---))))))))).))))))--...((((((...----((((((...(......)...))))))...))))))... ( -31.60) >DroYak_CAF1 14423 95 - 1 AACAGCUGAUUGCGACUGGCACACGGUUG---UUGCAGUCGUGCUGUU--UUUAUUUCUGUU----UUGUUGGAACUUCUUCGCUGCAGCGAUAAAGAGAGCGG .((.((((...(((((((.....))))))---)..)))).))(((.((--((((((.((((.----.((..((.....)).))..)))).)))))))).))).. ( -31.80) >consensus AACAGCUGAUUGCGACUGGCACACGGUUG___UUGCAGUCGUGCUGUU__UUUCUUUCUGUU____UUGUUGGAACUUUUUCGCUGCAGCGAUAAAGAGAGCGG ((((((((((((((((((.....)))))......))))))).))))))...........................((((.((((....)))).))))....... (-18.46 = -20.43 + 1.97)



| Location | 19,830,904 – 19,831,000 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19830904 96 + 23771897 CCAACAAAACAGAAAGAAAAACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUUUUGCCCGCUCACUCGCUCUGCUCUCUUGC .........((((....((((((((.((((((((...((....)).)).))))))......))))))))....((......))))))......... ( -21.60) >DroSec_CAF1 14582 96 + 1 CCAACAAAACAGAAAGAAAAACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUUUUGCGCGCUCACUCGCUCUGCUCUCUCGC .........((((..(.((((((((.((((((((...((....)).)).))))))......)))))))).)(((......)))))))......... ( -23.40) >DroSim_CAF1 17825 96 + 1 CCAACAAAACAGAAAGAAAAACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUUUUGCGCGCUCACUCGCUCUGCUCUCUCGC .........((((..(.((((((((.((((((((...((....)).)).))))))......)))))))).)(((......)))))))......... ( -23.40) >DroEre_CAF1 14542 92 + 1 UCAACAAAACAGAAAGAAAAACAGCACGACUGCAA---CAACCGUGUGCCAGUCGCAAUCAGCUGUU-UGCGCGCUCACUCGCUCCGCUCUUUCGC ...........((((((.(((((((.((((((..(---((....)))..))))))......))))))-)(((.((......))..))))))))).. ( -26.70) >DroYak_CAF1 14454 93 + 1 CCAACAAAACAGAAAUAAAAACAGCACGACUGCAA---CAACCGUGUGCCAGUCGCAAUCAGCUGUUUUAAACGCUCACUCGCUCUGUUCUUUUGC .(((.(.((((((....((((((((.((((((..(---((....)))..))))))......))))))))....((......)))))))).).))). ( -23.40) >consensus CCAACAAAACAGAAAGAAAAACAGCACGACUGCAACAACAACCGUGUGCCAGUCGCAAUCAGCUGUUUUGCGCGCUCACUCGCUCUGCUCUCUCGC .........((((..(.((((((((.((((((((...((....)).)).))))))......)))))))).)..((......))))))......... (-18.30 = -18.78 + 0.48)



| Location | 19,830,904 – 19,831,000 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -38.74 |
| Consensus MFE | -34.76 |
| Energy contribution | -34.48 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.85 |
| SVM RNA-class probability | 0.999956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19830904 96 - 23771897 GCAAGAGAGCAGAGCGAGUGAGCGGGCAAAACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUUUUUCUUUCUGUUUUGUUGG .(((.((((((((((......))(((.((((((((((((((((((.((((......)))))))))))))).)))))))).))))))))))).))). ( -40.40) >DroSec_CAF1 14582 96 - 1 GCGAGAGAGCAGAGCGAGUGAGCGCGCAAAACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUUUUUCUUUCUGUUUUGUUGG .(((.(((((((((((.(....).)))((((((((((((((((((.((((......)))))))))))))).))))))))....)))))))).))). ( -40.90) >DroSim_CAF1 17825 96 - 1 GCGAGAGAGCAGAGCGAGUGAGCGCGCAAAACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUUUUUCUUUCUGUUUUGUUGG .(((.(((((((((((.(....).)))((((((((((((((((((.((((......)))))))))))))).))))))))....)))))))).))). ( -40.90) >DroEre_CAF1 14542 92 - 1 GCGAAAGAGCGGAGCGAGUGAGCGCGCA-AACAGCUGAUUGCGACUGGCACACGGUUG---UUGCAGUCGUGCUGUUUUUCUUUCUGUUUUGUUGA .(((.(((((((((((.(....).)))(-((((((((((((((((.(((.....))))---))))))))).))))))).....)))))))).))). ( -35.90) >DroYak_CAF1 14454 93 - 1 GCAAAAGAACAGAGCGAGUGAGCGUUUAAAACAGCUGAUUGCGACUGGCACACGGUUG---UUGCAGUCGUGCUGUUUUUAUUUCUGUUUUGUUGG .(((.((((((((((......))....((((((((((((((((((.(((.....))))---))))))))).))))))))....)))))))).))). ( -35.60) >consensus GCGAGAGAGCAGAGCGAGUGAGCGCGCAAAACAGCUGAUUGCGACUGGCACACGGUUGUUGUUGCAGUCGUGCUGUUUUUCUUUCUGUUUUGUUGG .(((.((((((((((......))..(.((((((((((((((((((.(((.....)))...)))))))))).)))))))).)..)))))))).))). (-34.76 = -34.48 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:39 2006