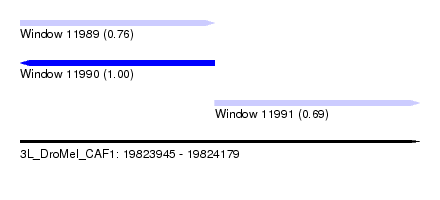
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,823,945 – 19,824,179 |
| Length | 234 |
| Max. P | 0.999569 |

| Location | 19,823,945 – 19,824,059 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.33 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -11.52 |
| Energy contribution | -14.00 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19823945 114 + 23771897 GAUUGGUAACAUUAAGCUGGCUGUAAACGAUGCAACGGCAACUAUACCGAAGUACAAGCAUAGAAAAUCGUUUAAGGAUC------AGCUCCAUCGUUUACAAACCACUUGAAUUAAGUU ...........(((((.(((.((((((((((((..(((........)))..))......................(((..------...)))))))))))))..))))))))........ ( -23.90) >DroSec_CAF1 7684 94 + 1 GAUUGGUAACAUGGAGCUGGUUGUAAACG--------------------AAGUACAAUCCUAGCAAAUCGGCCAAGGAGC------AGCUGCAUCGUUUACAAACAACUUGAAUUAAGUU ...........(.(((.((.(((((((((--------------------(.((((..((((.((......))..))))..------.).))).)))))))))).)).))).)........ ( -22.80) >DroSim_CAF1 10909 94 + 1 GAUUGGUAACAUGGAGCUGGUUGUAAACG--------------------AAGUACAAUCCUAGCAAAUCGCCCAAGGAGC------AGCUGCAUCGUUUACAAACAACUUGAAUUAAGUU ...........(.(((.((.(((((((((--------------------(.((((..((((.((.....))...))))..------.).))).)))))))))).)).))).)........ ( -22.20) >DroEre_CAF1 7623 99 + 1 GAUUGGUAACACCGAGCUGGUUGUAAACGAUGCAGCGGCAGCGAGUCCAAAG--------------------UCAGGAGCUUACAUCGC-UCAUCGUUUACAAACGAUGUGAAUUAGGUU ..((((.....))))((...((((((((((((.(((((..(..(((((....--------------------...))).))..).))))-))))))))))))).....)).......... ( -30.00) >DroYak_CAF1 7566 120 + 1 GAUUGGUAACAUCGAGCCGGUUGUGAACGAUGAAGCGGCAACAAGUCCGAAAUUGAAGCCUAAGAAAUCGGCUCAGGAGCUUAUAUCGCUUCAUCGUUUACAAACAACUUGAAUUAGGUU ...........(((((....(((((((((((((((((((..(((........)))..)))...((.((.((((....)))).)).))))))))))))))))))....)))))........ ( -42.10) >consensus GAUUGGUAACAUCGAGCUGGUUGUAAACGAUG_A_CGGCA_C_A__CC_AAGUACAAGCCUAGCAAAUCGGCUAAGGAGC______AGCUGCAUCGUUUACAAACAACUUGAAUUAAGUU ...........(((((.((.(((((((((((.....((........))...........................(((((.......)))))))))))))))).)).)))))........ (-11.52 = -14.00 + 2.48)



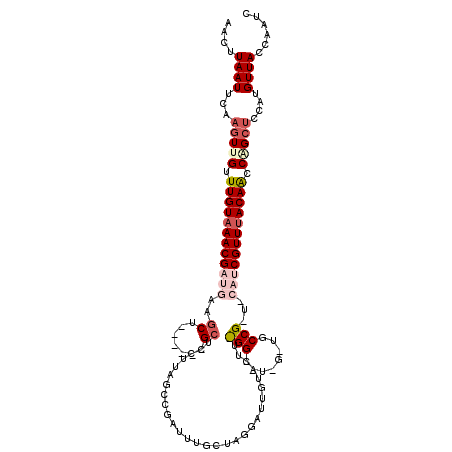
| Location | 19,823,945 – 19,824,059 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.33 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -11.62 |
| Energy contribution | -14.00 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.37 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19823945 114 - 23771897 AACUUAAUUCAAGUGGUUUGUAAACGAUGGAGCU------GAUCCUUAAACGAUUUUCUAUGCUUGUACUUCGGUAUAGUUGCCGUUGCAUCGUUUACAGCCAGCUUAAUGUUACCAAUC ....((((..((((((((.((((((((((.(((.------((((.......))))......((((((((....))))))..)).))).))))))))))))))).)))...))))...... ( -31.70) >DroSec_CAF1 7684 94 - 1 AACUUAAUUCAAGUUGUUUGUAAACGAUGCAGCU------GCUCCUUGGCCGAUUUGCUAGGAUUGUACUU--------------------CGUUUACAACCAGCUCCAUGUUACCAAUC ....((((...(((((.(((((((((((((((..------..((((.(((......))))))))))))..)--------------------))))))))).)))))....))))...... ( -25.60) >DroSim_CAF1 10909 94 - 1 AACUUAAUUCAAGUUGUUUGUAAACGAUGCAGCU------GCUCCUUGGGCGAUUUGCUAGGAUUGUACUU--------------------CGUUUACAACCAGCUCCAUGUUACCAAUC ....((((...(((((.(((((((((((((((..------..((((..(((.....))))))))))))..)--------------------))))))))).)))))....))))...... ( -24.70) >DroEre_CAF1 7623 99 - 1 AACCUAAUUCACAUCGUUUGUAAACGAUGA-GCGAUGUAAGCUCCUGA--------------------CUUUGGACUCGCUGCCGCUGCAUCGUUUACAACCAGCUCGGUGUUACCAAUC ..........((((((.(((((((((((((-(((..(..((.(((...--------------------....)))))..)...)))).))))))))))))......))))))........ ( -32.00) >DroYak_CAF1 7566 120 - 1 AACCUAAUUCAAGUUGUUUGUAAACGAUGAAGCGAUAUAAGCUCCUGAGCCGAUUUCUUAGGCUUCAAUUUCGGACUUGUUGCCGCUUCAUCGUUCACAACCGGCUCGAUGUUACCAAUC ....((((((.(((((.((((.((((((((((((..(((((.(((((((((.........))).))).....))))))))...)))))))))))).)))).))))).)).))))...... ( -41.00) >consensus AACUUAAUUCAAGUUGUUUGUAAACGAUGAAGCU______GCUCCUUAGCCGAUUUGCUAGGAUUGUACUU_GG__U_G_UGCCG_U_CAUCGUUUACAACCAGCUCCAUGUUACCAAUC ....((((...(((((.((((((((((((..((.......)).............................(((........)))...)))))))))))).)))))....))))...... (-11.62 = -14.00 + 2.38)


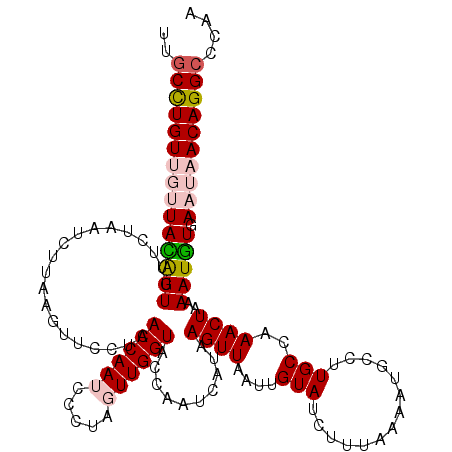
| Location | 19,824,059 – 19,824,179 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.94 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19824059 120 + 23771897 UUGCUUGUUGUUACAUUGUUUUAAUCUUAAGUUCCUAACCAAUCCCUAGUUGGUACCAAUCAUAAGUUAAUUGUAUCCUUAAAAUGCCUUGCCAAACUAAAAUGUGAAUAACAGGCUCAA ..(((((((((((((((...((((.((((.(......((((((.....))))))......).))))))))..((((.......)))).............)))))).))))))))).... ( -22.10) >DroSec_CAF1 7778 120 + 1 UUGCCUGUUGUUACAUUGUUUUUAUCGUAAGUUCCUAACCAAUCCCUAGUUGGUACCAAUCAUAAGUUAAUUGUAUCUUUAAAAUGCCUUGCCAAACUAAAAUGUUAAUAACAGGCCCAA ..(((((((((((((((.........(((((......((((((.....))))))..................((((.......)))))))))........)))).))))))))))).... ( -26.23) >DroSim_CAF1 11003 120 + 1 UUGCCUGUUGUUACAUUGUUGUAAUCGUAAGUUCCUAACCAAUCCCUAGUUGGUACCAAUCAUAAGUUAAUUGUAUCUUUAAAAUGCCUUGCCAAACUAAAAUGUUAAUAACAGGCUCAA ..(((((((((((((((...((....(((((......((((((.....))))))..................((((.......)))))))))...))...)))).))))))))))).... ( -27.70) >DroEre_CAF1 7722 116 + 1 UGACCUGU---UAUGUUGUUCUACUCUUAAGUUUCAAACCAAUACCUAUUUGGUACCAAUCAUAAGUUAAUUGUAUAU-UAAAAUGUCUUGCCAAACUGAAAUGUGAAUAACAGGCCGAA .(.(((((---(((((......))..(((.((((((.(((((.......)))))..(((.(((...(((((.....))-))).)))..)))......)))))).))))))))))).)... ( -18.50) >DroYak_CAF1 7686 117 + 1 UUGCCUGU---UAUAUUGUUCUUAUCUUAAGUUUCAAACCAAUACGUAUUUGGUACCAAUCAUAAGUUAAUUGUAUCUUUAAAAUGCCUUGCCAAACUAAAAUAUGAAUAACAGGCCGAA ..((((((---(((.(((..((((...))))...)))(((((.......))))).....(((((((((.(..((((.......))))..)....))))....)))))))))))))).... ( -20.10) >consensus UUGCCUGUUGUUACAUUGUUCUAAUCUUAAGUUCCUAACCAAUCCCUAGUUGGUACCAAUCAUAAGUUAAUUGUAUCUUUAAAAUGCCUUGCCAAACUAAAAUGUGAAUAACAGGCCCAA ..(((((((((((((((....................((((((.....))))))..........((((....(((..............)))..))))..))))).)))))))))).... (-16.86 = -17.94 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:33 2006