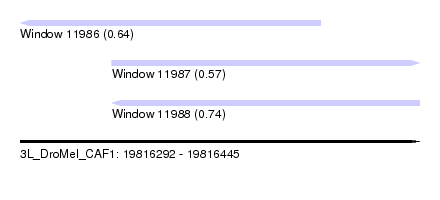
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,816,292 – 19,816,445 |
| Length | 153 |
| Max. P | 0.744685 |

| Location | 19,816,292 – 19,816,407 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -49.57 |
| Consensus MFE | -29.70 |
| Energy contribution | -30.57 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19816292 115 - 23771897 GGCUGGAUUCCAGGGCACAACAUUGCCAGCAUUCACGAGCGGACCGGCUGGCAACUUUGCCAACUUUCGGCCACAGUUCUCGGUGCCCGGCCAGCUAUCGAAUAAUUCCGGCG---UC-- ((((((...((.((((((....((((((((.(((......)))...))))))))....(((.......)))...........)))))))))))))).(((........)))..---..-- ( -40.70) >DroVir_CAF1 175 117 - 1 GGCCGGCUUUCAGGGCACCACCCUGCCGGCCUUCACGGGCGGACCCGCUGGCAACUUUGCCAACUUUCGGCCACAGUUCUCGGUGCCCGGGCAGCUGCCCAACAAUGCCG---GCACCUG .((((((.....(((((((...(((..((((....((((....)))).(((((....)))))......)))).))).....)))))))((((....))))......))))---))..... ( -59.30) >DroGri_CAF1 2 120 - 1 GGCCGGGUUUCAAGGCACCUCAUUGCCGGCAUUCACUGGCGGACCCGCCGGCAACUUUGCCAACUUUCGGCCACAGUUCUCGGUGCCCGGGCAGCUGGCCAACAACACCGGCGGCUCCUG .(((((.......((((......))))........)))))(((.(((((((((....)))).......(((((..((((((((...))))).)))))))).........))))).))).. ( -51.76) >DroWil_CAF1 6615 117 - 1 GGCUGGAUUCCAGGGCACAACUUUGCCGGCCUUUACCGGCGGACCGGCUGGCAAUUUUGCAAAUUUCCGACCACAGUUCUCUGUGCCCGGCCAGCUAUCGAAUAACUCGGGCA---AUUA ((((((...((.(((((((.....(((((......)))))((((.((.(((.((((.....)))).))).))...))))..))))))))))))))).((((.....))))...---.... ( -42.90) >DroMoj_CAF1 2 120 - 1 GGCCGGCUUUCAGGGCACCACCUUGCCGGCAUUCACGGGGGGACCAGCUGGGAACUUUGCCAAUUUUCGACCACAGUUCUCAGUGCCCGGGCAGCUGUCCAACAACGCCGGCGGCACCUG .((((((.....((((....((((.(((.......)))))))....((((((((((.((............)).))))))))))))))((((....))))......))))))........ ( -49.10) >DroAna_CAF1 2 117 - 1 GGCUGGAUUCCAGGGCACGACACUGCCCGCAUUCACCGGCGGACCGGCUGGCAACUUCGCCAGCUUCCGGCCACAGUUCUCGGUGCCCGGCCAGCUAUCGAAUAACGCCGGCG---CCUA (((((((.(((.(((((......)))))((........)))))..(((((((......)))))))))))))).........((((((.(((...............)))))))---)).. ( -53.66) >consensus GGCCGGAUUCCAGGGCACCACAUUGCCGGCAUUCACGGGCGGACCGGCUGGCAACUUUGCCAACUUUCGGCCACAGUUCUCGGUGCCCGGCCAGCUAUCCAACAACGCCGGCG___CCUG .((((((.....((((((......((((((.(((......)))...))))))((((..(((.......)))...))))....))))))((....))..........))))))........ (-29.70 = -30.57 + 0.86)

| Location | 19,816,327 – 19,816,445 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -24.25 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19816327 118 + 23771897 AGAACUGUGGCCGAAAGUUGGCAAAGUUGCCAGCCGGUCCGCUCGUGAAUGCUGGCAAUGUUGUGCCCUGGAAUCCAGCCGGGAAUUGUGCCAUCAAUUGAGCUG--CAACAGAAAUAAA ....(((((((((...(((((((....)))))))))))).(((((....((.(((((.....((..(((((.......))))).))..))))).))..)))))..--..))))....... ( -41.10) >DroVir_CAF1 212 114 + 1 AGAACUGUGGCCGAAAGUUGGCAAAGUUGCCAGCGGGUCCGCCCGUGAAGGCCGGCAGGGUGGUGCCCUGAAAGCCGGCCGGAAACUGUGCCAUCAAUUGAGCUG--CAAAA----CCAA .....((..(((((..(.(((((.((((.((.(((((....)))))...(((((((((((.....))))....))))))))).)))).))))).)..))).))..--))...----.... ( -54.30) >DroGri_CAF1 42 118 + 1 AGAACUGUGGCCGAAAGUUGGCAAAGUUGCCGGCGGGUCCGCCAGUGAAUGCCGGCAAUGAGGUGCCUUGAAACCCGGCCGGGAAUUGAGGCAUCAAUUGAGCUG--CAAUCAAACCAAA .((..((..(((((..(((((((...(..(.((((....)))).)..).))))))).....(((((((..(..(((....)))..)..)))))))..))).))..--)).))........ ( -50.20) >DroWil_CAF1 6652 120 + 1 AGAACUGUGGUCGGAAAUUUGCAAAAUUGCCAGCCGGUCCGCCGGUAAAGGCCGGCAAAGUUGUGCCCUGGAAUCCAGCCGGGAAUUGAGCCAUCAGUUGUGCUAAUGAACAGAACAAGA ....((((((((((......(((....)))...)))).))((((((....))))))......(..((((((.......)))))((((((....)))))))..)......))))....... ( -36.40) >DroMoj_CAF1 42 118 + 1 AGAACUGUGGUCGAAAAUUGGCAAAGUUCCCAGCUGGUCCCCCCGUGAAUGCCGGCAAGGUGGUGCCCUGAAAGCCGGCCGGAAAUUGUGCCAUCAAUUGAGCUG--CAAAAUUGACAAA .....((..(((((....(((((...((((..((.((....)).))....((((((.(((......)))....)))))).))))....)))))....)))).)..--))........... ( -35.50) >DroAna_CAF1 39 118 + 1 AGAACUGUGGCCGGAAGCUGGCGAAGUUGCCAGCCGGUCCGCCGGUGAAUGCGGGCAGUGUCGUGCCCUGGAAUCCAGCCGGAAAUUGUGCCAUUAAUUGAGCUG--CAAAAUAAAAAUU .....((..((((((.(((((((....)))))))...))))(((((......(((((......)))))(((...))))))))...................))..--))........... ( -40.80) >consensus AGAACUGUGGCCGAAAGUUGGCAAAGUUGCCAGCCGGUCCGCCCGUGAAUGCCGGCAAUGUGGUGCCCUGAAAUCCAGCCGGAAAUUGUGCCAUCAAUUGAGCUG__CAAAAGAAACAAA ........((((((....(((((.((((.((.((.((...((........)).((((......)))).......)).)).)).)))).)))))....))).)))................ (-24.25 = -24.28 + 0.03)


| Location | 19,816,327 – 19,816,445 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -25.44 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19816327 118 - 23771897 UUUAUUUCUGUUG--CAGCUCAAUUGAUGGCACAAUUCCCGGCUGGAUUCCAGGGCACAACAUUGCCAGCAUUCACGAGCGGACCGGCUGGCAACUUUGCCAACUUUCGGCCACAGUUCU .......((((.(--(...((....))(((((......((((((((..(((..((((......)))).((........)))))))))))))......))))).......)).)))).... ( -36.70) >DroVir_CAF1 212 114 - 1 UUGG----UUUUG--CAGCUCAAUUGAUGGCACAGUUUCCGGCCGGCUUUCAGGGCACCACCCUGCCGGCCUUCACGGGCGGACCCGCUGGCAACUUUGCCAACUUUCGGCCACAGUUCU ..((----..(((--..(((.((..(.(((((.((((.(((((((((....((((.....)))))))))))......((((....)))))).)))).))))).).)).)))..)))..)) ( -45.30) >DroGri_CAF1 42 118 - 1 UUUGGUUUGAUUG--CAGCUCAAUUGAUGCCUCAAUUCCCGGCCGGGUUUCAAGGCACCUCAUUGCCGGCAUUCACUGGCGGACCCGCCGGCAACUUUGCCAACUUUCGGCCACAGUUCU ...((...(((((--..((((....)).))..))))).))(((((((......((((......))))((((....((((((....))))))......))))....)))))))........ ( -41.00) >DroWil_CAF1 6652 120 - 1 UCUUGUUCUGUUCAUUAGCACAACUGAUGGCUCAAUUCCCGGCUGGAUUCCAGGGCACAACUUUGCCGGCCUUUACCGGCGGACCGGCUGGCAAUUUUGCAAAUUUCCGACCACAGUUCU ....(..((((.((((((.....))))))((..((((.((((((((..(((..((((......)))).(((......)))))))))))))).))))..))............))))..). ( -40.10) >DroMoj_CAF1 42 118 - 1 UUUGUCAAUUUUG--CAGCUCAAUUGAUGGCACAAUUUCCGGCCGGCUUUCAGGGCACCACCUUGCCGGCAUUCACGGGGGGACCAGCUGGGAACUUUGCCAAUUUUCGACCACAGUUCU ...(((((((...--......)))))))((((.(.((((((((((((....(((......))).)))))).......((....))...)))))).).))))................... ( -35.60) >DroAna_CAF1 39 118 - 1 AAUUUUUAUUUUG--CAGCUCAAUUAAUGGCACAAUUUCCGGCUGGAUUCCAGGGCACGACACUGCCCGCAUUCACCGGCGGACCGGCUGGCAACUUCGCCAGCUUCCGGCCACAGUUCU .............--............((((.......((((.(((((....(((((......)))))..)))))))))((((..(((((((......)))))))))))))))....... ( -42.70) >consensus UUUGUUUAUUUUG__CAGCUCAAUUGAUGGCACAAUUCCCGGCCGGAUUCCAGGGCACCACAUUGCCGGCAUUCACGGGCGGACCGGCUGGCAACUUUGCCAACUUUCGGCCACAGUUCU .....................(((((..((........))(((((((......((((.....((((((((.(((......)))...))))))))...))))....))))))).))))).. (-25.44 = -26.38 + 0.95)

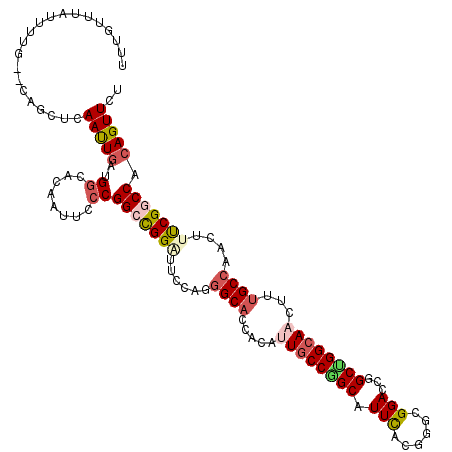
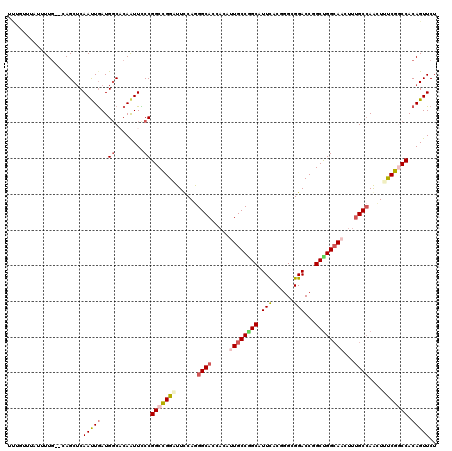
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:31 2006