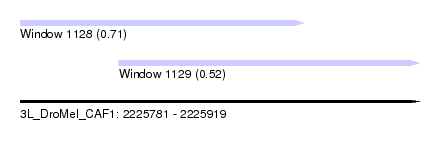
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,225,781 – 2,225,919 |
| Length | 138 |
| Max. P | 0.711939 |

| Location | 2,225,781 – 2,225,879 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -14.41 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
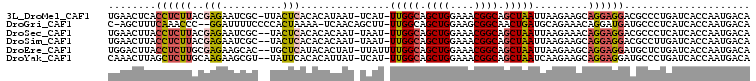
>3L_DroMel_CAF1 2225781 98 + 23771897 GUUCAUUGCCAAAGUGGAUUACAAGUAACUACAGUGAACUCACCUCUUACGAGAAUCGC-UUACUCACACAUAAU-UCAU-UUGGCAGCUGGAAACGGCAG ......(((((((.(((((((...((((.....(((....)))...))))(((......-...))).....))))-))))-))))))((((....)))).. ( -27.30) >DroVir_CAF1 6461 96 + 1 GUUCAUAGCCAAAGUGGAUUACAAGUAAGUCGUUU-AGCAUCUAUGUUC--AGAUUUGCCUGUCUAAAA-GCAACAGCAA-UUGGCAGCUGGAAGCGGCAA .......(((((..(((((.....(((((((....-((((....)))).--.)))))))..)))))...-((....))..-))))).((((....)))).. ( -24.70) >DroSec_CAF1 6718 96 + 1 GUUCAUUGCCAAAGUGGAUUACAAGUAACUA-AGUGAACUUACCUCUUACGAGAAUCGC--UACUCACACACAAU-UAAU-UUGGCAGCUGGAAACGGCAG ......(((((((...((((....((((.((-((....))))....))))(((......--..)))......)))-)..)-))))))((((....)))).. ( -25.20) >DroSim_CAF1 6814 97 + 1 GUUCAUUGCCAAAGUGGAUUACAAGUAACUACAGUGAACUUACCUCUUACGAGAAUCGC--UACUCACACACAAU-UAAU-UUGGCAGCUGGAAACGGCAG ......(((((((((((.(((....)))))))(((((......(((....)))..))))--).............-...)-))))))((((....)))).. ( -25.70) >DroEre_CAF1 14511 98 + 1 GUUCAUUGCCAAAGUGGAUUACAAGUAAUUACAGUGGACUUACCUCUUGCGAGAAGCAC--UGCUCAUACACUAU-UUAUUUUGGCAGCUGGAAACGGCAG ......(((((((((((((.....(((....(((((..(((..(((....)))))))))--))....)))...))-)))))))))))((((....)))).. ( -30.60) >DroYak_CAF1 6976 97 + 1 GUUCAUUGCCAAAGUGGAUUACAAGUAAUAACUGCAAACUUAGCUCUUGCAAGAAGCGU--UAUUCACACAUUAU-UCAU-UUGGCAGCUGGAAACGGCAG ......(((((((.(((((.....((((((((.((...(((.((....)))))..))))--)))).)).....))-))))-))))))((((....)))).. ( -29.50) >consensus GUUCAUUGCCAAAGUGGAUUACAAGUAACUACAGUGAACUUACCUCUUACGAGAAUCGC__UACUCACACACAAU_UCAU_UUGGCAGCUGGAAACGGCAG ......((((((.(((.................((((.........))))(((..........)))...........))).))))))((((....)))).. (-14.41 = -15.17 + 0.75)

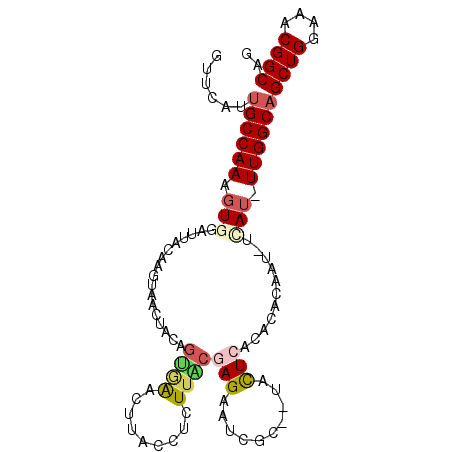
| Location | 2,225,815 – 2,225,919 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2225815 104 + 23771897 UGAACUCACCUCUUACGAGAAUCGC-UUACUCACACAUAAU-UCAU-UUGGCAGCUGGAAACGGCAGCUAAUUAAGAAGCAGGAGGACGCCCUGAUCACCAAUGACA ........((((((.((((......-...)))........(-((..-(((((.((((....)))).)))))....)))).))))))..................... ( -23.40) >DroGri_CAF1 6508 102 + 1 C-AGCUUUCAAACCC--GGAUUUUCCCCACUAAAA-UCAACAGCUU-UUGGCAGCUGGAAGCGGCAACUGAUGCAGAAACAGGAUGAUGCCCUCAUCACCAAUGACA .-.((..(((...((--(((((((.......))))-))..(((((.-.....)))))....)))....))).)).......((.(((((....)))))))....... ( -21.40) >DroSec_CAF1 6751 103 + 1 UGAACUUACCUCUUACGAGAAUCGC--UACUCACACACAAU-UAAU-UUGGCAGCUGGAAACGGCAGCUAAUUAAGAAACAGGAGGACGCCCUCAUCACCAAUGACA ........((((((..(((......--..)))........(-((((-(.(((.((((....)))).))))))))).....))))))......((((.....)))).. ( -24.30) >DroSim_CAF1 6848 103 + 1 UGAACUUACCUCUUACGAGAAUCGC--UACUCACACACAAU-UAAU-UUGGCAGCUGGAAACGGCAGCUAAUUAAGAAGCAGGAGGACGCCUUGAUCACCAAUGACA ........((((((.((((......--..)))........(-((((-(.(((.((((....)))).)))))))))...).))))))..................... ( -24.30) >DroEre_CAF1 14545 104 + 1 UGGACUUACCUCUUGCGAGAAGCAC--UGCUCAUACACUAU-UUAUUUUGGCAGCUGGAAACGGCAGCUAAUUAAGAAGCAGGAGGAUGCUCUGAUCACCAAUGACA .(((....(((((((((((......--..)))........(-(((..(((((.((((....)))).))))).))))..))))))))....))).............. ( -31.10) >DroYak_CAF1 7010 103 + 1 CAAACUUAGCUCUUGCAAGAAGCGU--UAUUCACACAUUAU-UCAU-UUGGCAGCUGGAAACGGCAGCUAAUCAAGAAGCAGGAGGAUGCCCUGAUCACCAAUGACA .........(((((((..(((....--..)))........(-((..-(((((.((((....)))).)))))....))))))))))(((......))).......... ( -26.80) >consensus UGAACUUACCUCUUACGAGAAUCGC__UACUCACACACAAU_UCAU_UUGGCAGCUGGAAACGGCAGCUAAUUAAGAAGCAGGAGGACGCCCUGAUCACCAAUGACA ........((((((..(((..........)))...............(((((.((((....)))).))))).........))))))..................... (-16.16 = -16.50 + 0.34)
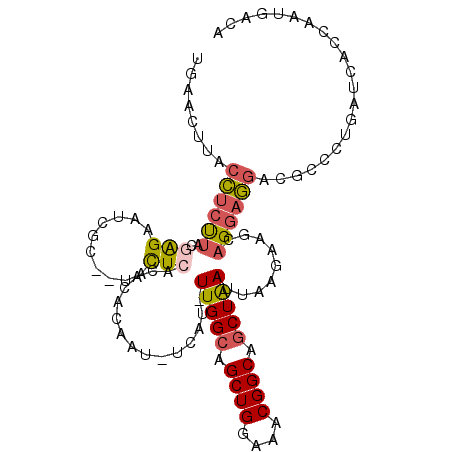
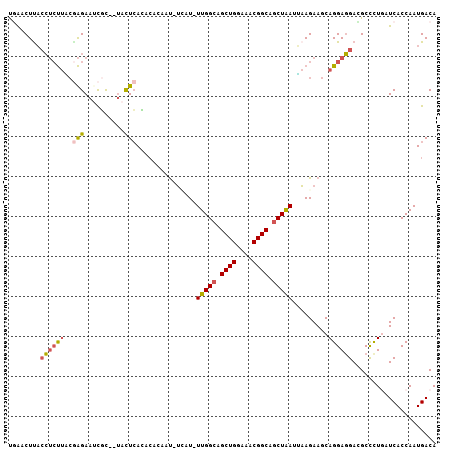
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:50 2006