| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
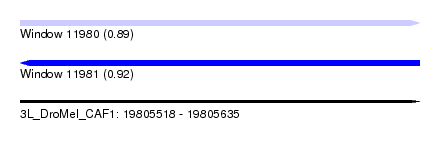
| Location | 19,805,518 – 19,805,635 |
| Length | 117 |
| Max. P | 0.915160 |

| Location | 19,805,518 – 19,805,635 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -29.87 |
| Energy contribution | -29.67 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19805518 117 + 23771897 UUAUCUACUGCCAUUCCAGUGUGGCUUCUUUAUUGCAUGUG--UUUUCAUGUGCAUGGGUAUUUGCGUCUCCGCCGCCAAUGACGUCACAAUAGAAAAGCUAACCGAAUGGGGUGG-CAG .......(((((((((((...((((((.(((((((((((..--(......)..))))(((....(((....))).)))..........))))))).))))))......))))))))-))) ( -41.00) >DroSec_CAF1 14114 108 + 1 UUAUCUACUGCCAUUCCAGUGUGGCUUCUUUAU------------UUCAUGUGCAUGGGUAUUUGCGUCUCCGCCGCCAGUGACGUCACAAUAGAAAAGCUCACCGAAUGGGGUGGCCAG .......(((((((((((..(((((((.(((((------------(..((((.(((.(((....(((....))).))).)))))))...)))))).)))).)))....)))))))).))) ( -34.90) >DroSim_CAF1 35099 108 + 1 UUAUCUACUGCCAUUCCAGUGUGGCUUCUUUAU------------UUCAUGUGCAUGGGUAUUUGCGUCUCCGCCGCCAGUGACGUCACAAUAGAAAAGCUCACCGAAUGGGGUGGCCAG .......(((((((((((..(((((((.(((((------------(..((((.(((.(((....(((....))).))).)))))))...)))))).)))).)))....)))))))).))) ( -34.90) >DroEre_CAF1 7888 120 + 1 UUAUCUGCUGCCAUUCCAGUGUGGCUUCUUUAUUUCAUGUGCAUUUACAUGUGCAUGGGUAUUUGCGUCUCCGCCGCCAGUGACGUCACAAUAGAAAAGCUCAUCGAAUGGGGUGGGCAG ....((((..((((((((..(((((((.((((((.((((..(((....)))..))))(((....(((....))).))).(((....))))))))).)))).)))....)))))))))))) ( -40.30) >DroYak_CAF1 16476 120 + 1 UUAUCUGCUGCCAUUCCAGUGUGGCUUCUUUAUUUCAUGUGUGUUUUCAUGUGCAUGGGUAUUUGCGUCUCCGCCGCCAGUGACGUCACAAUAGAAAAGCUCAUCGACUGGGGUGGGCAG ....((((..(((((((((((((((((.((((((.((((..(((....)))..))))(((....(((....))).))).(((....))))))))).)))).)))..)))))))))))))) ( -40.60) >consensus UUAUCUACUGCCAUUCCAGUGUGGCUUCUUUAUU_CAUGUG__UUUUCAUGUGCAUGGGUAUUUGCGUCUCCGCCGCCAGUGACGUCACAAUAGAAAAGCUCACCGAAUGGGGUGGCCAG .......(((((((((((..(((((((.(((((................((((.(((..((((.(((.......))).)))).)))))))))))).)))).)))....)))))))).))) (-29.87 = -29.67 + -0.20)


| Location | 19,805,518 – 19,805,635 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -27.15 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
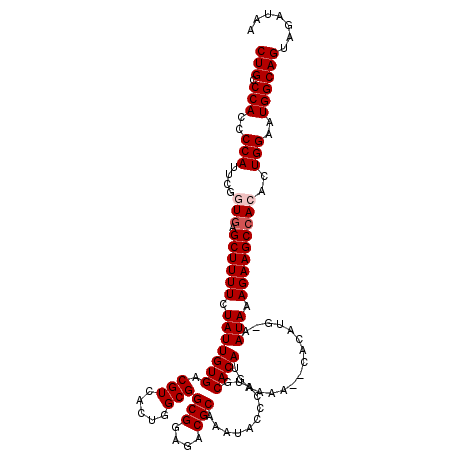
>3L_DroMel_CAF1 19805518 117 - 23771897 CUG-CCACCCCAUUCGGUUAGCUUUUCUAUUGUGACGUCAUUGGCGGCGGAGACGCAAAUACCCAUGCACAUGAAAA--CACAUGCAAUAAAGAAGCCACACUGGAAUGGCAGUAGAUAA (((-(((..(((....((..((((((.((((((...(((((.((..(((....))).....)).))).))(((....--..))))))))).)))))).))..)))..))))))....... ( -34.60) >DroSec_CAF1 14114 108 - 1 CUGGCCACCCCAUUCGGUGAGCUUUUCUAUUGUGACGUCACUGGCGGCGGAGACGCAAAUACCCAUGCACAUGAA------------AUAAAGAAGCCACACUGGAAUGGCAGUAGAUAA ...((((..(((....(((.((((((.(((((((.(((.....)))(((....)))...........))))....------------))).)))))))))..)))..))))......... ( -33.10) >DroSim_CAF1 35099 108 - 1 CUGGCCACCCCAUUCGGUGAGCUUUUCUAUUGUGACGUCACUGGCGGCGGAGACGCAAAUACCCAUGCACAUGAA------------AUAAAGAAGCCACACUGGAAUGGCAGUAGAUAA ...((((..(((....(((.((((((.(((((((.(((.....)))(((....)))...........))))....------------))).)))))))))..)))..))))......... ( -33.10) >DroEre_CAF1 7888 120 - 1 CUGCCCACCCCAUUCGAUGAGCUUUUCUAUUGUGACGUCACUGGCGGCGGAGACGCAAAUACCCAUGCACAUGUAAAUGCACAUGAAAUAAAGAAGCCACACUGGAAUGGCAGCAGAUAA ((((.....((((((.(((.((((((.(((((((.(((.....)))(((....)))...))).((((..(((....)))..)))).)))).))))))))...).))))))..)))).... ( -33.80) >DroYak_CAF1 16476 120 - 1 CUGCCCACCCCAGUCGAUGAGCUUUUCUAUUGUGACGUCACUGGCGGCGGAGACGCAAAUACCCAUGCACAUGAAAACACACAUGAAAUAAAGAAGCCACACUGGAAUGGCAGCAGAUAA (((((((..(((((.(.((.((((((.(((((((.(((.....)))(((....))).......)))...((((........)))).)))).))))))))))))))..)))..)))).... ( -36.70) >consensus CUGCCCACCCCAUUCGGUGAGCUUUUCUAUUGUGACGUCACUGGCGGCGGAGACGCAAAUACCCAUGCACAUGAAAA__CACAUG_AAUAAAGAAGCCACACUGGAAUGGCAGUAGAUAA (((.(((..(((....(((.((((((.(((((((.(((.....)))(((....)))...........))))................))).)))))))))..)))..))))))....... (-27.15 = -27.75 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:23 2006