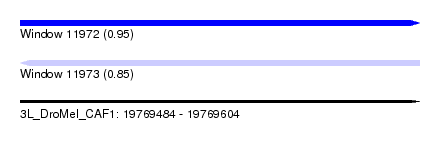
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,769,484 – 19,769,604 |
| Length | 120 |
| Max. P | 0.950753 |

| Location | 19,769,484 – 19,769,604 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -19.23 |
| Energy contribution | -20.26 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
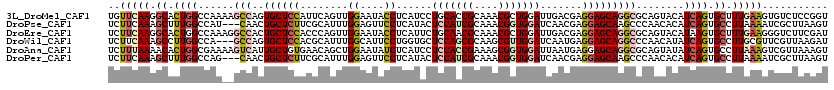
>3L_DroMel_CAF1 19769484 120 + 23771897 UGUUCAAGGCACUGGCCAAAAGCCAGUGCUCCAUUCAGUUGGAAUACCUCAUCCUGCACCGCAAACGCUGGAUUGACGAGGAGCAGGCGCAGUACAUCAGUGCUUUGAAGUGUCUCCGGU ..(((((((((((((......(((..((((((((((.....)))).(.((((((.((.........)).))).))).).)))))))))........)))))))))))))........... ( -40.24) >DroPse_CAF1 15109 117 + 1 UCUUCAAAGCUUUGGCCAU---CAACUGCUCUUCGCAUUUGGAGUUCCUCAUACUCCAUCGCAAACGGUGGAUCAACGAGGAGCAAGCCCAACACAUCAGUGCCUUAAAAUCGCUUAAGU ......((((...(((..(---(((.(((.....))).)))).(((((((....(((((((....))))))).....)))))))..)))...(((....)))..........)))).... ( -33.70) >DroEre_CAF1 14044 120 + 1 UCUUCAAGGCACUGGCCAAAGGCCACUGCUCCACCCAGUUGGAAUACCUCAUUCUGCAACGCAAACGCUGGAUUGACGAGGAGCAGGCGCAGUACAUAAGUGCUUUGAAGGGUCUUCGAU (((((((((((((((((...)))).(((((((..((....))....(.(((.(((((.........)).))).))).).)))))))............)))))))))))))......... ( -44.40) >DroWil_CAF1 18632 117 + 1 UCUUCAAAGCCUUGGCCA---GCCAGUGCUCCACGCAUUUGGCAUUCCUGGUGCUCCAGCGCAAGCGUUGGAUCAAUGAGGAGCAGGCCCAACAUAUCAGUGCCUUGCGUUCGUUAAGAU ..........((((((..---((((((((.....))).)))))..((((.((..(((((((....)))))))...)).))))((((((.(.........).))).)))....)))))).. ( -39.30) >DroAna_CAF1 13409 120 + 1 UCUUUAAAACACUGGCGAAAAGUCAUUGCUGUGAACAGCUGGAAUAUCUCAUCCUCCACCGAAAGCGGUGGAUUAAUGAGGAGCAGGCGCAGUAUAUCAGUGCCUUAAAGUCGUUAAAGU .((((((...(((.((......((...((((....))))..))...((((((..(((((((....)))))))...)))))).))((((((.........))))))...)))..)))))). ( -39.80) >DroPer_CAF1 15059 117 + 1 UCUUCAAAGCUUUGGCCAG---CAACUGCUCUUCGCAUUUGGAGUUCCUCAUACUCCAUCGCAAACGGUGGAUCAACGAGGAGCAAGCCCAACACAUCAGUGCCUUAAAAUCGCUUAAGU ......((((...(((...---(((.(((.....))).)))..(((((((....(((((((....))))))).....)))))))..)))...(((....)))..........)))).... ( -33.10) >consensus UCUUCAAAGCACUGGCCAA__GCCACUGCUCCACGCAGUUGGAAUACCUCAUACUCCACCGCAAACGGUGGAUCAACGAGGAGCAGGCCCAACACAUCAGUGCCUUAAAGUCGCUUAAGU ..(((((.((.((((......(((..((((((........((....))......(((((((....))))))).......)))))))))........)))).)).)))))........... (-19.23 = -20.26 + 1.03)
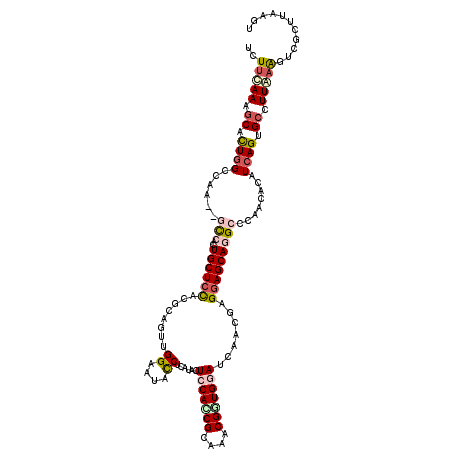
| Location | 19,769,484 – 19,769,604 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.97 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19769484 120 - 23771897 ACCGGAGACACUUCAAAGCACUGAUGUACUGCGCCUGCUCCUCGUCAAUCCAGCGUUUGCGGUGCAGGAUGAGGUAUUCCAACUGAAUGGAGCACUGGCUUUUGGCCAGUGCCUUGAACA ...(....)((((((..(((....))).(((((((.((....(((.......)))...)))))))))..))))))..((((......))))((((((((.....))))))))........ ( -42.80) >DroPse_CAF1 15109 117 - 1 ACUUAAGCGAUUUUAAGGCACUGAUGUGUUGGGCUUGCUCCUCGUUGAUCCACCGUUUGCGAUGGAGUAUGAGGAACUCCAAAUGCGAAGAGCAGUUG---AUGGCCAAAGCUUUGAAGA .(((((((........(((((....))))).((((...(((((((...((((.((....)).))))..)))))))....(((.(((.....))).)))---..))))...))))..))). ( -36.40) >DroEre_CAF1 14044 120 - 1 AUCGAAGACCCUUCAAAGCACUUAUGUACUGCGCCUGCUCCUCGUCAAUCCAGCGUUUGCGUUGCAGAAUGAGGUAUUCCAACUGGGUGGAGCAGUGGCCUUUGGCCAGUGCCUUGAAGA ..........((((((.(((((.....(((((.((....((((((.....(((((....)))))....))))))...(((....))).)).)))))((((...))))))))).)))))). ( -43.20) >DroWil_CAF1 18632 117 - 1 AUCUUAACGAACGCAAGGCACUGAUAUGUUGGGCCUGCUCCUCAUUGAUCCAACGCUUGCGCUGGAGCACCAGGAAUGCCAAAUGCGUGGAGCACUGGC---UGGCCAAGGCUUUGAAGA .((((.....(((((((((.(..(....)..)))))((((((...((.((((.((....)).)))).))..))))..))....)))))(((((..(((.---...)))..))))).)))) ( -36.00) >DroAna_CAF1 13409 120 - 1 ACUUUAACGACUUUAAGGCACUGAUAUACUGCGCCUGCUCCUCAUUAAUCCACCGCUUUCGGUGGAGGAUGAGAUAUUCCAGCUGUUCACAGCAAUGACUUUUCGCCAGUGUUUUAAAGA ..........(((((((((((((.......(((.......((((((..(((((((....))))))).))))))........((((....))))..........)))))))))))))))). ( -40.51) >DroPer_CAF1 15059 117 - 1 ACUUAAGCGAUUUUAAGGCACUGAUGUGUUGGGCUUGCUCCUCGUUGAUCCACCGUUUGCGAUGGAGUAUGAGGAACUCCAAAUGCGAAGAGCAGUUG---CUGGCCAAAGCUUUGAAGA .(((((((........(((((....))))).((((.(((((((((...((((.((....)).))))..)))))))........(((.....)))...)---).))))...))))..))). ( -38.00) >consensus ACCUAAGCGACUUCAAGGCACUGAUGUACUGCGCCUGCUCCUCGUUAAUCCACCGUUUGCGAUGGAGGAUGAGGAAUUCCAAAUGCGUAGAGCAGUGGC__UUGGCCAAUGCUUUGAAGA ..........(((((((((((....)).(((.(((....((((((...((((.((....)).))))..)))))).........(((.....))).........)))))).))))))))). (-21.80 = -22.97 + 1.17)

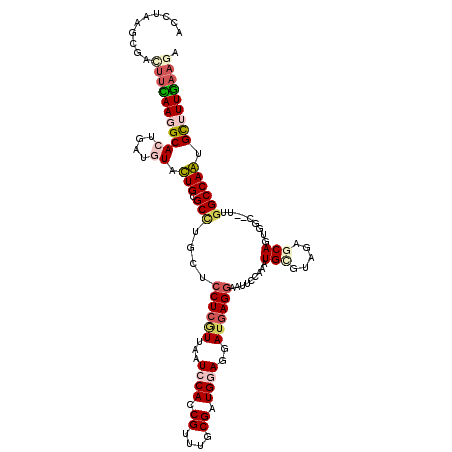
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:15 2006