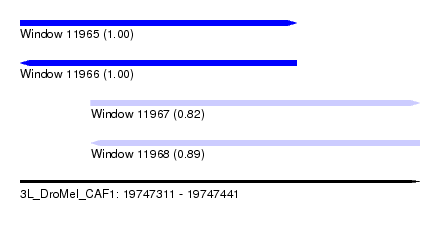
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,747,311 – 19,747,441 |
| Length | 130 |
| Max. P | 0.999490 |

| Location | 19,747,311 – 19,747,401 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19747311 90 + 23771897 GUGCAAGCAAA-AAAGUUGAGUGAAACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAACACACACAUGUCCGUACAUAUGU ((((..(((..-....(((((...........((((((......)))))))))))((((....)))).........)))..))))...... ( -20.10) >DroSec_CAF1 18133 89 + 1 GUGCAAGCAAA-AAAGUCGAGGGAAACGAAAAA-GGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUACACACAUGUCGGUACAUAUGU ((((..(((..-......((((....)......-((((......))))..)))..((((....)))).........)))..))))...... ( -21.50) >DroSim_CAF1 18181 90 + 1 GUGCAAGCAAA-AAAGUCGAGGGAAACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUACACACAUGUCGGUACAUAUGU ((((..(((..-......((((....).....((((((......)))))))))..((((....)))).........)))..))))...... ( -24.10) >DroEre_CAF1 18456 89 + 1 GUGCAAGCAAAAAAAGUCGAGUGAAACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAGUCAAAUACACACAUGUCGGUACAUAU-- ((((..(((......(((((.(((........((((((......)))))).))).)))))..((.....)).....)))..))))....-- ( -22.40) >DroYak_CAF1 18050 88 + 1 GUGCAAGCAAAAAAAGUCGAGUGAAACGAA-AACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUACACACAUGUCGGUACAUAU-- ((((..(((.........(((.........-.((((((......)))))))))..((((....)))).........)))..))))....-- ( -20.50) >consensus GUGCAAGCAAA_AAAGUCGAGUGAAACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUACACACAUGUCGGUACAUAUGU ((((..(((.........(((...........((((((......)))))))))..((((....)))).........)))..))))...... (-19.02 = -19.22 + 0.20)

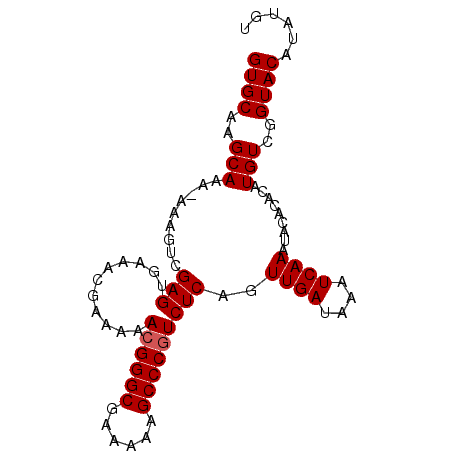
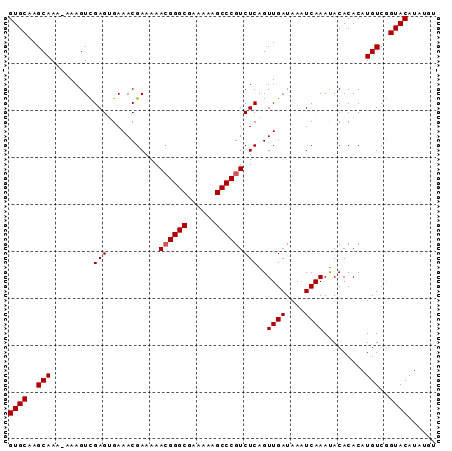
| Location | 19,747,311 – 19,747,401 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19747311 90 - 23771897 ACAUAUGUACGGACAUGUGUGUGUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUUUCACUCAACUUU-UUUGCUUGCAC ((((((((....))))))))((((..(...........(((((((((......)))))))))..(((......)))...-....)..)))) ( -23.40) >DroSec_CAF1 18133 89 - 1 ACAUAUGUACCGACAUGUGUGUAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCC-UUUUUCGUUUCCCUCGACUUU-UUUGCUUGCAC ((((((((....))))))))....((((......(((.((((.((((......))))-.)))).)))....))))....-........... ( -17.80) >DroSim_CAF1 18181 90 - 1 ACAUAUGUACCGACAUGUGUGUAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUUUCCCUCGACUUU-UUUGCUUGCAC ((((((((....))))))))(((...............(((((((((......)))))))))(((.......)))....-......))).. ( -22.40) >DroEre_CAF1 18456 89 - 1 --AUAUGUACCGACAUGUGUGUAUUUGACUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUUUCACUCGACUUUUUUUGCUUGCAC --((((((....))))))(((((...............(((((((((......)))))))))(((.......)))...........))))) ( -20.70) >DroYak_CAF1 18050 88 - 1 --AUAUGUACCGACAUGUGUGUAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUU-UUCGUUUCACUCGACUUUUUUUGCUUGCAC --((((((....))))))(((((...((....))(((.(((((((((......)))))))-)).)))...................))))) ( -21.00) >consensus ACAUAUGUACCGACAUGUGUGUAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUUUCACUCGACUUU_UUUGCUUGCAC ..((((((....))))))(((((.((((....))))..(((((((((......)))))))))........................))))) (-18.74 = -18.98 + 0.24)

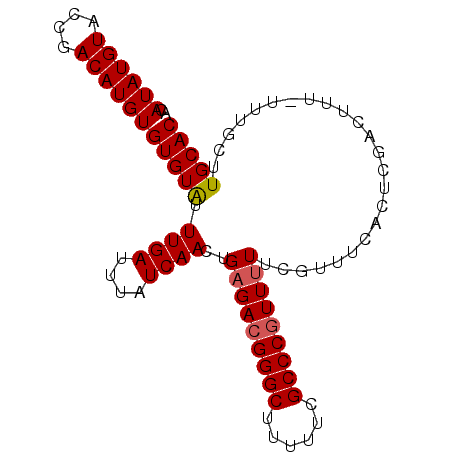
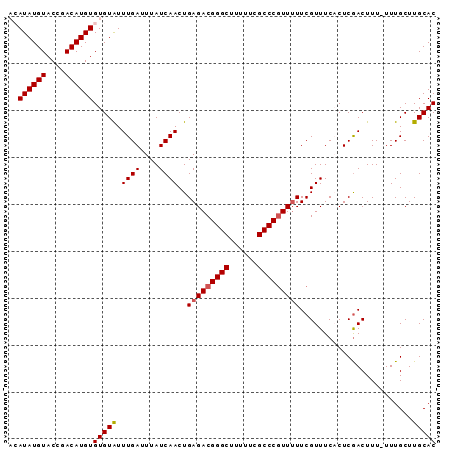
| Location | 19,747,334 – 19,747,441 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -13.17 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19747334 107 + 23771897 AACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAACA------CACACAUGUCCGUACAUAUGUACAUAUGUACACAUGAUCCACACCAUUUACCACAAGUAGU ...((...((((((......)))))).))..((((....))))...------....(((((..((((((((....)))))))))))))......................... ( -25.00) >DroSec_CAF1 18156 106 + 1 AACGAAAAA-GGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUA------CACACAUGUCGGUACAUAUGUACCUAUGUACACAUGAUAAACACCAUUUACCACAAGUUGU (((((....-((((......))))(((......)))...)).....------....(((((..(((((((......))))))))))))....................))).. ( -21.50) >DroSim_CAF1 18204 104 + 1 AACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUA------CACACAUGUCGGUACAUAUGUACCUAUGUACACAUGAUCCACACCAUUUA---CAAGUAGU ...((...((((((......)))))).))..((((....))))...------....(((((..(((((((......))))))))))))..............---........ ( -23.20) >DroEre_CAF1 18480 99 + 1 AACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAGUCAAAUA------CACACAUGUCGGUACAUAU--------GUACACAUGAUCCACACCAUUUACCAUAAGUAGU ...((...((((((......)))))).)).((.(((..((.....)------)...(((((..((((....--------)))))))))))).))................... ( -20.40) >DroYak_CAF1 18074 98 + 1 AACGAA-AACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUA------CACACAUGUCGGUACAUAU--------GUACACAUGAUCCACACCAUUUACCAUAAGUAGU (((...-.((((((......))))))....))).............------....(((((..((((....--------)))))))))......................... ( -20.50) >DroAna_CAF1 18193 100 + 1 AACAAAAAUCAGGCGAAAAAGCCCGUCUCACUUGAUAAACCAAAUAAACCGACACACAUGUCGGCCCAGGUACAUAUAACUACACAU------------AUA-AGUAAGUAGU .......((((((.((...........)).))))))..(((.......((((((....))))))....))).......(((((((..------------...-.))..))))) ( -16.90) >consensus AACGAAAAACGGGCGAAAAAGCCCGUCUCAGUUGAUAAAUCAAAUA______CACACAUGUCGGUACAUAUGUAC_UAUGUACACAUGAUCCACACCAUUUACCACAAGUAGU ..(((...((((((......)))))).....)))......................(((((..((((............)))))))))......................... (-13.17 = -14.20 + 1.03)

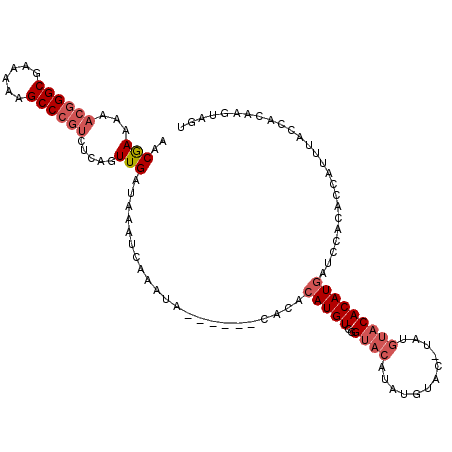
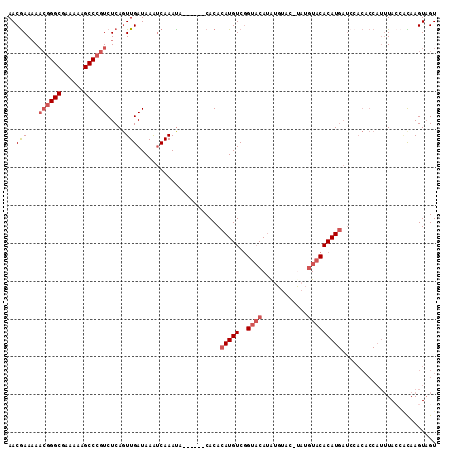
| Location | 19,747,334 – 19,747,441 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -16.65 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19747334 107 - 23771897 ACUACUUGUGGUAAAUGGUGUGGAUCAUGUGUACAUAUGUACAUAUGUACGGACAUGUGUG------UGUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUU ........((((((((.........(((((((((....)))))))))..(((((((....)------))))))))))))))...(((((((((......)))))))))..... ( -35.20) >DroSec_CAF1 18156 106 - 1 ACAACUUGUGGUAAAUGGUGUUUAUCAUGUGUACAUAGGUACAUAUGUACCGACAUGUGUG------UAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCC-UUUUUCGUU ........((((((((.(.((....(((((((.....(((((....))))).)))))))..------.)).).))))))))((.((((.((((......))))-.)))).)). ( -28.50) >DroSim_CAF1 18204 104 - 1 ACUACUUG---UAAAUGGUGUGGAUCAUGUGUACAUAGGUACAUAUGUACCGACAUGUGUG------UAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUU ........---.....(((((((((((((..(((((.(((((....)))))...)))))..------))..))))))))..)))(((((((((......)))))))))..... ( -32.90) >DroEre_CAF1 18480 99 - 1 ACUACUUAUGGUAAAUGGUGUGGAUCAUGUGUAC--------AUAUGUACCGACAUGUGUG------UAUUUGACUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUU .((((((((.....)))).)))).(((.((((((--------((((((....)))))))))------))).)))......(((.(((((((((......)))))))))..))) ( -30.10) >DroYak_CAF1 18074 98 - 1 ACUACUUAUGGUAAAUGGUGUGGAUCAUGUGUAC--------AUAUGUACCGACAUGUGUG------UAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUU-UUCGUU ..(((.....)))......((((((((.((((((--------((((((....)))))))))------))).)))))))).(((.(((((((((......)))))))-)).))) ( -32.80) >DroAna_CAF1 18193 100 - 1 ACUACUUACU-UAU------------AUGUGUAGUUAUAUGUACCUGGGCCGACAUGUGUGUCGGUUUAUUUGGUUUAUCAAGUGAGACGGGCUUUUUCGCCUGAUUUUUGUU ....((((((-(((------------(((((....)))))))(((((((((((((....))))))))))...))).....))))))).(((((......)))))......... ( -31.20) >consensus ACUACUUAUGGUAAAUGGUGUGGAUCAUGUGUACAUA_GUACAUAUGUACCGACAUGUGUG______UAUUUGAUUUAUCAACUGAGACGGGCUUUUUCGCCCGUUUUUCGUU ...................((((((((...(((......((((((((......))))))))......))).)))))))).....(((((((((......)))))))))..... (-16.65 = -19.40 + 2.75)

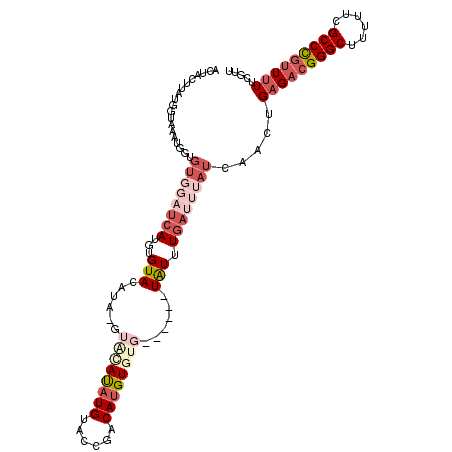
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:10 2006