
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,731,486 – 19,731,594 |
| Length | 108 |
| Max. P | 0.903331 |

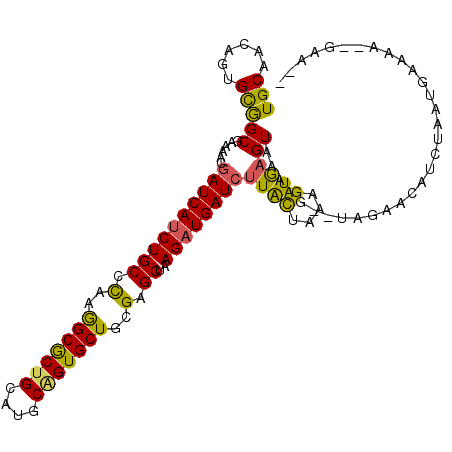
| Location | 19,731,486 – 19,731,594 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -23.37 |
| Energy contribution | -22.57 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19731486 108 + 23771897 UGCAACAGUGCGGCGAAAACGAUCAUCUGCCCAAGGCGCUGCAUGCUGUGCUGCGAGCUAAGAUGAUCUUACUAAAGUGAGUAAAAC--UAGAACAACUAAUGAAAA--GAA-- ......(((...((......(((((((((((...)))((((((.((...))))).)))..))))))))((((....))))))...))--).................--...-- ( -24.90) >DroVir_CAF1 2647 110 + 1 UGCAGCAGAGCAGCGACAAUGAUCAUCUGCCUAAAGCACUGCAUGCGGUGCUGCGUGCCAAGAUGAUAUUGCUAAUGUAUGUGGUUGAUUCGAACAUC---UGCAUAUAUUUC- (((((..(((((((.(((...(((((((((....(((((((....)))))))....))..)))))))..(((....)))))).)))).)))......)---))))........- ( -35.10) >DroSec_CAF1 2737 105 + 1 UGCAACAGUGCGGCGAAAACGAUCAUCUGCCCAAGGCGCUGCAUGCAGUGCUGCGAGCUAAGAUGAUCUUACUAAAGUGAGUGAACG--UAGGA---UUAAUGAAAA--GAA-- .(((....))).(((.....((((((((((.(..(((((((....)))))))..).))..))))))))(((((......))))).))--)....---..........--...-- ( -30.60) >DroSim_CAF1 2735 108 + 1 UGCAACAGUGCGGCGAAAACGAUCAUCUGCCCAAGGCGCUGCAUGCAGUGCUGCGAGCUAAGAUGAUCUUACUAAAGUGAGUAUACG--UAGGACCAUUAAUGAAAA--GAA-- .......((.(.(((.....((((((((((.(..(((((((....)))))))..).))..)))))))).((((......))))..))--).).))............--...-- ( -30.10) >DroEre_CAF1 2852 110 + 1 UGCAAAAGUGUGGCGAAAACGAUCAACUGCCUAAGGCGCUGCAUGCAGUGCUACGGGCUAAGAUGAUCUUAUUAAAGUGAGUAAAAC--UAGAACAUCCGAUGAAAA--AUUCA ......(((...((......(((((.((((((..(((((((....)))))))..))))..)).)))))((((....))))))...))--).................--..... ( -26.10) >DroYak_CAF1 2726 107 + 1 UGCAAAAGAGUGGCGAAAACGAUCAUCUGCCCAAGGCGCUGCACGCAGUGCUACGAGCUAAGAUGAUCUUACUAAAGUAAGUAAAAG--UAGAGCAUCAAAUGUAAA--AA--- ((((...((...((......((((((((((.(..(((((((....)))))))..).))..))))))))(((((......)))))...--....)).))...))))..--..--- ( -30.00) >consensus UGCAACAGUGCGGCGAAAACGAUCAUCUGCCCAAGGCGCUGCAUGCAGUGCUGCGAGCUAAGAUGAUCUUACUAAAGUGAGUAAAAG__UAGAACAUCUAAUGAAAA__GAA__ (((......)))((......((((((((((.(..(((((((....)))))))..).))..))))))))((((....))))))................................ (-23.37 = -22.57 + -0.80)


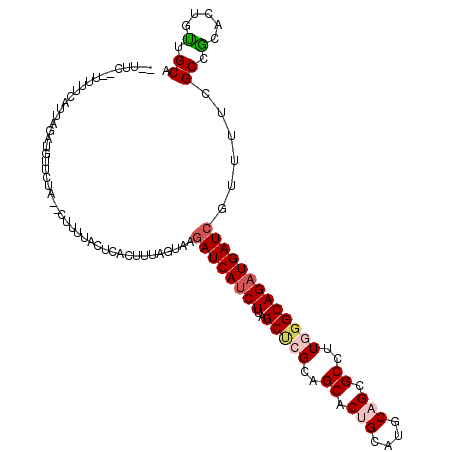
| Location | 19,731,486 – 19,731,594 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19731486 108 - 23771897 --UUC--UUUUCAUUAGUUGUUCUA--GUUUUACUCACUUUAGUAAGAUCAUCUUAGCUCGCAGCACAGCAUGCAGCGCCUUGGGCAGAUGAUCGUUUUCGCCGCACUGUUGCA --...--.....(((((.....)))--)).(((((......)))))((((((((..(((.(((((...)).))))))(((...))))))))))).........(((....))). ( -25.90) >DroVir_CAF1 2647 110 - 1 -GAAAUAUAUGCA---GAUGUUCGAAUCAACCACAUACAUUAGCAAUAUCAUCUUGGCACGCAGCACCGCAUGCAGUGCUUUAGGCAGAUGAUCAUUGUCGCUGCUCUGCUGCA -........(((.---(((((...............))))).)))...............((((((..(((.((..(((.....)))((..(...)..)))))))..)))))). ( -26.26) >DroSec_CAF1 2737 105 - 1 --UUC--UUUUCAUUAA---UCCUA--CGUUCACUCACUUUAGUAAGAUCAUCUUAGCUCGCAGCACUGCAUGCAGCGCCUUGGGCAGAUGAUCGUUUUCGCCGCACUGUUGCA --...--..........---.....--...................((((((((..(((((..((.(((....))).))..))))))))))))).........(((....))). ( -25.90) >DroSim_CAF1 2735 108 - 1 --UUC--UUUUCAUUAAUGGUCCUA--CGUAUACUCACUUUAGUAAGAUCAUCUUAGCUCGCAGCACUGCAUGCAGCGCCUUGGGCAGAUGAUCGUUUUCGCCGCACUGUUGCA --...--...........(((...(--(...((((......)))).((((((((..(((((..((.(((....))).))..)))))))))))))))....)))(((....))). ( -30.40) >DroEre_CAF1 2852 110 - 1 UGAAU--UUUUCAUCGGAUGUUCUA--GUUUUACUCACUUUAAUAAGAUCAUCUUAGCCCGUAGCACUGCAUGCAGCGCCUUAGGCAGUUGAUCGUUUUCGCCACACUUUUGCA .(((.--...(((.((((((.(((.--(((...........))).))).)))))..(((.(..((.(((....))).))..).))).).))).....))).............. ( -20.30) >DroYak_CAF1 2726 107 - 1 ---UU--UUUACAUUUGAUGCUCUA--CUUUUACUUACUUUAGUAAGAUCAUCUUAGCUCGUAGCACUGCGUGCAGCGCCUUGGGCAGAUGAUCGUUUUCGCCACUCUUUUGCA ---..--...........(((....--...(((((......)))))((((((((..(((((..((.(((....))).))..))))))))))))).................))) ( -28.90) >consensus __UUC__UUUUCAUUAGAUGUUCUA__CUUUUACUCACUUUAGUAAGAUCAUCUUAGCUCGCAGCACUGCAUGCAGCGCCUUGGGCAGAUGAUCGUUUUCGCCGCACUGUUGCA ..............................................((((((((..(((((..((.(((....))).))..)))))))))))))......((.(.....).)). (-19.30 = -20.00 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:03 2006