
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,725,883 – 19,726,037 |
| Length | 154 |
| Max. P | 0.883931 |

| Location | 19,725,883 – 19,726,001 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -26.40 |
| Energy contribution | -27.08 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
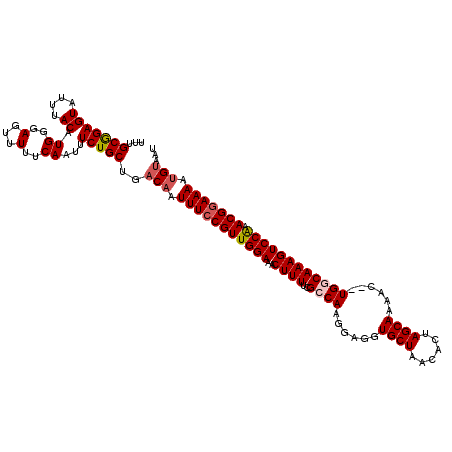
>3L_DroMel_CAF1 19725883 118 + 23771897 UUUGCGGAGUAUUUACAUGGGAGUUUUUCAUUUUCUGCUGACAAUUUCCGUUGGAAACUUUUCGCCAAGGAGGUGCUAACACUAGCAAAAC--UGGCAAAGUCCAAACGGAAAAUGUAAU ...(((((((....))(((..(...)..)))..)))))..(((.(((((((((((..((((..((((.(....(((((....)))))...)--))))))))))).)))))))).)))... ( -33.00) >DroSec_CAF1 35205 118 + 1 UUUGCGGAGUAUUUACAUGGGAGUUUUUCAAUUUCUGCUAACAAUUUCCGUUGGAAACUUUUCGCCAAGGAGGUGCUAACACUAGCAAAAC--UGGCAAAGUCCAAACGGAAAAUGUAAU ...(((((((....)).((..(...)..))...)))))..(((.(((((((((((..((((..((((.(....(((((....)))))...)--))))))))))).)))))))).)))... ( -32.50) >DroSim_CAF1 37376 118 + 1 UUUGCGGAGUAUUUACAUGGGAGUUUUUCAAUUUCUGCUGACAAUUUCCGUUGGAAACUUUUCGCCAAGGAGGUGCUAACACUAGCAAAAC--UGGCAAAGUCCAAACGGAAAAUGUAAU ...(((((((....)).((..(...)..))...)))))..(((.(((((((((((..((((..((((.(....(((((....)))))...)--))))))))))).)))))))).)))... ( -32.60) >DroEre_CAF1 37959 120 + 1 UUUGCGGAGUAUUUACAUGGGAGUUUUUCAAUUUCUGCUGACAAUUUCCGUUGGAAACUUUUCGCCAAGAAGGUGCUAGACUAAGCAAAUCUCUGUCAAAGUCCGAACGAAAAAAGUAAU (((((((((.((((....(((((((....)))))))(((.....(((((...)))))((...((((.....))))..))....)))))))))))).)))).................... ( -21.60) >DroYak_CAF1 36037 118 + 1 UUUGCAGAGUAUUUACAUGGGAGUUUUUCAAUUUCUGCUGACAAUUUCCGUUGGAAUCUUUUCGCCAAGAAGGUGCUAAACUAAGCAAAUC--UGCCAAAGUCCAAACGGAAAAUGAAAU ...(((((((....)).((..(...)..))...)))))...((.(((((((((((..((((..((..(((...((((......))))..))--))).))))))).)))))))).)).... ( -27.70) >consensus UUUGCGGAGUAUUUACAUGGGAGUUUUUCAAUUUCUGCUGACAAUUUCCGUUGGAAACUUUUCGCCAAGGAGGUGCUAACACUAGCAAAAC__UGGCAAAGUCCAAACGGAAAAUGUAAU ...(((((((....)).((..(...)..))...)))))..(((.(((((((((((..((((..((((......((((......))))......)))))))))))).))))))).)))... (-26.40 = -27.08 + 0.68)

| Location | 19,725,923 – 19,726,037 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19725923 114 - 23771897 CUCUCUGUGCUUCUGUUUCUCCGGCUUUCUGCAUA----AAUUACAUUUUCCGUUUGGACUUUGCCA--GUUUUGCUAGUGUUAGCACCUCCUUGGCGAAAAGUUUCCAACGGAAAUUGU .....(((((....(((.....))).....)))))----....(((.((((((((.(((.(((((((--(...(((((....))))).....)))))))).....))))))))))).))) ( -30.10) >DroSec_CAF1 35245 114 - 1 CUCUCUGUUCUUCUGUUUCUCCGGCUUACUGCAUA----AAUUACAUUUUCCGUUUGGACUUUGCCA--GUUUUGCUAGUGUUAGCACCUCCUUGGCGAAAAGUUUCCAACGGAAAUUGU .......................((.....))...----....(((.((((((((.(((.(((((((--(...(((((....))))).....)))))))).....))))))))))).))) ( -27.30) >DroSim_CAF1 37416 114 - 1 CUCUCUGUGCUUCUGUUUCUCCGGCUUACUGCAUA----AAUUACAUUUUCCGUUUGGACUUUGCCA--GUUUUGCUAGUGUUAGCACCUCCUUGGCGAAAAGUUUCCAACGGAAAUUGU .....(((((....(((.....))).....)))))----....(((.((((((((.(((.(((((((--(...(((((....))))).....)))))))).....))))))))))).))) ( -30.10) >DroEre_CAF1 37999 116 - 1 CUGUAUGUGUUUCUGUUUCUGCGUCCCACUGCAUC----AAUUACUUUUUUCGUUCGGACUUUGACAGAGAUUUGCUUAGUCUAGCACCUUCUUGGCGAAAAGUUUCCAACGGAAAUUGU .((((.(((....(((....)))...)))))))..----....((..((((((((.(((((((....((((..((((......))))...)))).....))))..)))))))))))..)) ( -24.30) >DroYak_CAF1 36077 118 - 1 AUCUCGGUGUUUCUGUUUCUGCGGCCUACUGCAUAUGCAUAUUUCAUUUUCCGUUUGGACUUUGGCA--GAUUUGCUUAGUUUAGCACCUUCUUGGCGAAAAGAUUCCAACGGAAAUUGU .............(((...(((((....)))))...))).....((.((((((((.(((((((.(((--((..((((......))))...)))..))..))))..))))))))))).)). ( -28.80) >consensus CUCUCUGUGCUUCUGUUUCUCCGGCUUACUGCAUA____AAUUACAUUUUCCGUUUGGACUUUGCCA__GUUUUGCUAGUGUUAGCACCUCCUUGGCGAAAAGUUUCCAACGGAAAUUGU ......((((..(((......)))......)))).........(((.((((((((.(((.(((((((......((((......))))......))))))).....))))))))))).))) (-21.88 = -22.68 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:58 2006