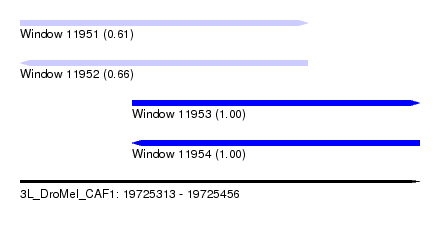
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,725,313 – 19,725,456 |
| Length | 143 |
| Max. P | 0.999962 |

| Location | 19,725,313 – 19,725,416 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 99.35 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -34.37 |
| Energy contribution | -34.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
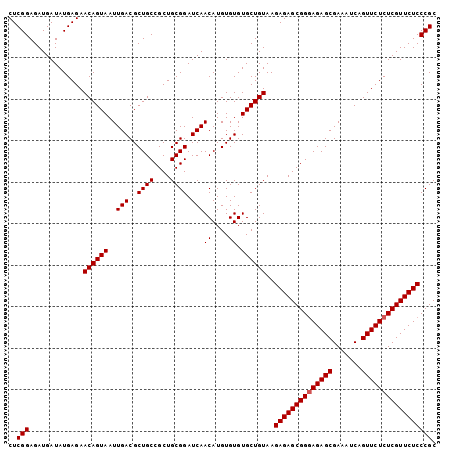
>3L_DroMel_CAF1 19725313 103 + 23771897 CUCGGAGAUGAUAUGAGAACAGUAAUUGACGCUGCCGCUGCGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCUCUCGUUCUCCCGC ..(((.............((((((..(((..((((....)))).)))((....)).))))))..((((((((((((((.......))))))))))))))))). ( -36.20) >DroSec_CAF1 34647 103 + 1 CUCGGAGAUGAUAUGAGAACAGUAAUUGACGCUGCCGCUGCGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGUAGAGCGAAAUCAGUUCUCUCGUUCUCCCGC ...((((((((...((((((.(...(((...((((((((((((....(((....))).)))).....))))))))..)))...).))))))))).)))))... ( -33.80) >DroSim_CAF1 36799 103 + 1 CUCGGAGAUGAUAUGAGAACAGUAAUUGACGCUGCCGCUGCGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCUCUCGUUCUCCCGC ..(((.............((((((..(((..((((....)))).)))((....)).))))))..((((((((((((((.......))))))))))))))))). ( -36.20) >consensus CUCGGAGAUGAUAUGAGAACAGUAAUUGACGCUGCCGCUGCGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCUCUCGUUCUCCCGC ..(((.............((((((..(((..((((....)))).)))((....)).))))))..((((((((((((((.......))))))))))))))))). (-34.37 = -34.70 + 0.33)


| Location | 19,725,313 – 19,725,416 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 99.35 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
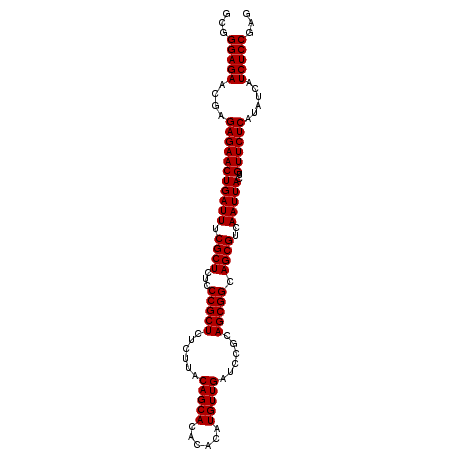
>3L_DroMel_CAF1 19725313 103 - 23771897 GCGGGAGAACGAGAGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCGCAGCGGCAGCGUCAAUUACUGUUCUCAUAUCAUCUCCGAG ...(((((....(((((((((((.((((...(((((......(((((......)))))......))))).))))..)))))..))))))......)))))... ( -30.50) >DroSec_CAF1 34647 103 - 1 GCGGGAGAACGAGAGAACUGAUUUCGCUCUACCGCUCUCUUACAGCACACACAUGUUGAUCCGCAGCGGCAGCGUCAAUUACUGUUCUCAUAUCAUCUCCGAG ...(((((....(((((((((((.((((...(((((......(((((......)))))......))))).))))..)))))..))))))......)))))... ( -30.50) >DroSim_CAF1 36799 103 - 1 GCGGGAGAACGAGAGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCGCAGCGGCAGCGUCAAUUACUGUUCUCAUAUCAUCUCCGAG ...(((((....(((((((((((.((((...(((((......(((((......)))))......))))).))))..)))))..))))))......)))))... ( -30.50) >consensus GCGGGAGAACGAGAGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCGCAGCGGCAGCGUCAAUUACUGUUCUCAUAUCAUCUCCGAG ...(((((....(((((((((((.((((...(((((......(((((......)))))......))))).))))..)))))..))))))......)))))... (-30.50 = -30.50 + 0.00)


| Location | 19,725,353 – 19,725,456 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -37.63 |
| Energy contribution | -37.71 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
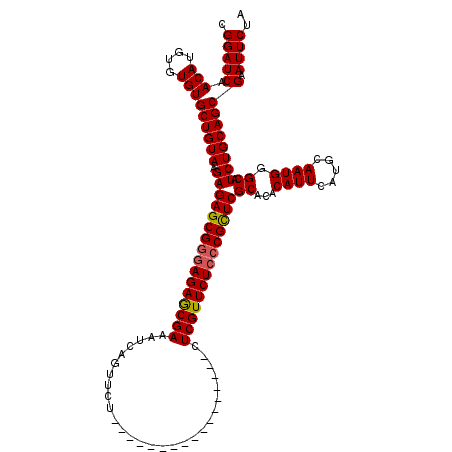
>3L_DroMel_CAF1 19725353 103 + 23771897 CGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCU-----------------CUCGUUCUCCCGCUCGCACACAUUCAUGCAAUGGGCAUCUGCAGCGAAUUCUA .(((((.(((....)))((((((.((((((((((((((((..........-----------------.))))))))))))))((...((((.....)))).)).))))))))).)))).. ( -41.70) >DroSec_CAF1 34687 103 + 1 CGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGUAGAGCGAAAUCAGUUCU-----------------CUCGUUCUCCCGCUCGCACACAUUCAUGCAAUGGGCAUCUGCAGCGAAUUCUA .(((......((((((((..((..(((((((..(((((.......)))))-----------------..)))))))..)).))))))))(((.((((.........))))..))).))). ( -35.00) >DroSim_CAF1 36839 103 + 1 CGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCU-----------------CUCGUUCUCCCGCUCGCACACAUUCAUGCAAUGGGCAUCUGCAGCGAAUUCUA .(((((.(((....)))((((((.((((((((((((((((..........-----------------.))))))))))))))((...((((.....)))).)).))))))))).)))).. ( -41.70) >DroEre_CAF1 37418 120 + 1 CGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAACGAAAUCAGCGGGAGAACGAGAGAACUGAUUUCGUUCUCCCGCUCGCACACAUUCAUGCAAUGGGCAUCUGCAGCGAAUUCUA .(((((.(((....)))((((((.(((((((((((((((((((((((....)..(....)..))))))))))))))))))))((...((((.....)))).)).))))))))).)))).. ( -53.20) >DroYak_CAF1 35529 102 + 1 CGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCG-----------------CUCGUUCUC-CGUUCGCACACAUUCAUGCAAUGGGCAUCUGCAGCGAAUUCUA .(((((..(((((((((((.....(((((((((.((((.......)))).-----------------)))))))))-.....)))))))..))))......((....))...).)))).. ( -35.10) >consensus CGGAUCAACAUGUGUGUGCUGUAAGAGAGCGGGAGAGCGAAAUCAGUUCU_________________CUCGUUCUCCCGCUCGCACACAUUCAUGCAAUGGGCAUCUGCAGCGAAUUCUA .(((((.(((....)))((((((.((((((((((((((((............................))))))))))))))((...((((.....)))).)).))))))))).)))).. (-37.63 = -37.71 + 0.08)

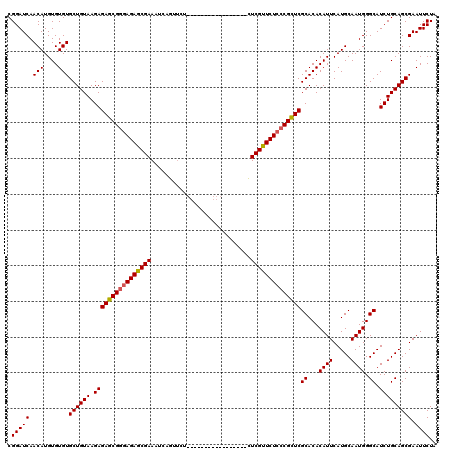
| Location | 19,725,353 – 19,725,456 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -38.94 |
| Consensus MFE | -31.80 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19725353 103 - 23771897 UAGAAUUCGCUGCAGAUGCCCAUUGCAUGAAUGUGUGCGAGCGGGAGAACGAG-----------------AGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCG ....(((((.(((((.......))))))))))((((((((((((((((.((((-----------------(.......))))).)))))))))).......))))))............. ( -39.71) >DroSec_CAF1 34687 103 - 1 UAGAAUUCGCUGCAGAUGCCCAUUGCAUGAAUGUGUGCGAGCGGGAGAACGAG-----------------AGAACUGAUUUCGCUCUACCGCUCUCUUACAGCACACACAUGUUGAUCCG ....(((((.(((((.......))))))))))((((((((((((.(((.((((-----------------(.......))))).))).)))))).......))))))............. ( -32.91) >DroSim_CAF1 36839 103 - 1 UAGAAUUCGCUGCAGAUGCCCAUUGCAUGAAUGUGUGCGAGCGGGAGAACGAG-----------------AGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCG ....(((((.(((((.......))))))))))((((((((((((((((.((((-----------------(.......))))).)))))))))).......))))))............. ( -39.71) >DroEre_CAF1 37418 120 - 1 UAGAAUUCGCUGCAGAUGCCCAUUGCAUGAAUGUGUGCGAGCGGGAGAACGAAAUCAGUUCUCUCGUUCUCCCGCUGAUUUCGUUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCG ....(((((.(((((.......))))))))))(((((((((((((((((((((((((((..............))))))))))))))))))))).......))))))............. ( -52.05) >DroYak_CAF1 35529 102 - 1 UAGAAUUCGCUGCAGAUGCCCAUUGCAUGAAUGUGUGCGAACG-GAGAACGAG-----------------CGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCG ....(((((.(((((.......))))))))))((((((.((.(-(((...(((-----------------((((.....))))))).....)))).))...))))))............. ( -30.30) >consensus UAGAAUUCGCUGCAGAUGCCCAUUGCAUGAAUGUGUGCGAGCGGGAGAACGAG_________________AGAACUGAUUUCGCUCUCCCGCUCUCUUACAGCACACACAUGUUGAUCCG ....(((((.(((((.......))))))))))((((((((((((((((.((((..........................)))).)))))))))).......))))))............. (-31.80 = -32.24 + 0.44)

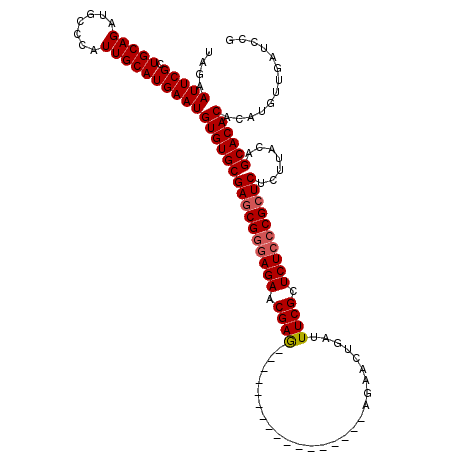

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:56 2006