
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,724,709 – 19,724,962 |
| Length | 253 |
| Max. P | 0.999357 |

| Location | 19,724,709 – 19,724,824 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19724709 115 + 23771897 CUUU-UUACUU--UUUUCACCUUUUUCGCACUCUUUGGGUUUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAU--AUGUAUGCAUAAUAUCUUUUUCUGUGAG ....-......--............((((.((..(((((((..((....))...)))))))..))(((((.....((((((..(((((..--..)))))..))))))...))))))))). ( -20.30) >DroSec_CAF1 34024 120 + 1 CUUUUUUACUUUUUUUUCACCUUUUUCGCACUCUUUGGGUUUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAUAUAUGUAUGCAUAAUAUCGUUUUCUGUGAG .........................((((.((..(((((((..((....))...)))))))..))(((((.....((((((..((((((....))))))..))))))...))))))))). ( -20.90) >DroSim_CAF1 36176 119 + 1 CUUUUUUACUUU-UUUUCACCUUUUUCGCACUCUUUGGGUUUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAUAUAUGUAUGCAUAAUAUCGUUUUCUGUGAG ............-............((((.((..(((((((..((....))...)))))))..))(((((.....((((((..((((((....))))))..))))))...))))))))). ( -20.90) >DroEre_CAF1 36886 114 + 1 CUUU-UUACUUU-UUUUCACCUUUUUCGCACUC--UGGGUUUUGUUUUUGCCCCAAACCAAUCAGAGAAAAAUAUAUAUUAAACAUACAU--AUGUAUGCAUAGUAUCGUUUUCUGUGAG ....-.......-....................--.((((.........))))........(((.((((((....((((((..(((((..--..)))))..))))))..)))))).))). ( -20.40) >DroYak_CAF1 34899 115 + 1 CUUU-UUACUUC-UUUUCACCUU-UUCGCACUCUAUGGGUUUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAU--AUAUAUGCAUAGUAUCGUUUUCUGUGAG ....-.......-..........-.(((((((((.((((((..((....))...))))))...))))...(((((((((.........))--)))))))...............))))). ( -15.70) >consensus CUUU_UUACUUU_UUUUCACCUUUUUCGCACUCUUUGGGUUUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAU__AUGUAUGCAUAAUAUCGUUUUCUGUGAG ....................................((((.........))))........(((.(((((.....((((((..((((((....))))))..))))))...))))).))). (-18.44 = -18.40 + -0.04)



| Location | 19,724,709 – 19,724,824 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19724709 115 - 23771897 CUCACAGAAAAAGAUAUUAUGCAUACAU--AUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAAACCCAAAGAGUGCGAAAAAGGUGAAAA--AAGUAA-AAAG (((.((((.(((.((((((.((((((..--..)))))).)))))).....))).))))......(((((............))))).))).................--......-.... ( -22.80) >DroSec_CAF1 34024 120 - 1 CUCACAGAAAACGAUAUUAUGCAUACAUAUAUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAAACCCAAAGAGUGCGAAAAAGGUGAAAAAAAAGUAAAAAAG .(((((((((...((((((.(((((((....))))))).)))))).....))))).........(((((............)))))..............))))................ ( -22.70) >DroSim_CAF1 36176 119 - 1 CUCACAGAAAACGAUAUUAUGCAUACAUAUAUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAAACCCAAAGAGUGCGAAAAAGGUGAAAA-AAAGUAAAAAAG .(((((((((...((((((.(((((((....))))))).)))))).....))))).........(((((............)))))..............))))...-............ ( -22.70) >DroEre_CAF1 36886 114 - 1 CUCACAGAAAACGAUACUAUGCAUACAU--AUGUAUGUUUAAUAUAUAUUUUUCUCUGAUUGGUUUGGGGCAAAAACAAAACCCA--GAGUGCGAAAAAGGUGAAAA-AAAGUAA-AAAG .((((((((((..(((.((.((((((..--..)))))).)).)))....))))))......(.((((((............))))--)).).........))))...-.......-.... ( -20.80) >DroYak_CAF1 34899 115 - 1 CUCACAGAAAACGAUACUAUGCAUAUAU--AUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAAACCCAUAGAGUGCGAA-AAGGUGAAAA-GAAGUAA-AAAG .((((......((.((((......((((--((((((.....))))))))))..............((((............))))...))))))..-...))))...-.......-.... ( -18.50) >consensus CUCACAGAAAACGAUAUUAUGCAUACAU__AUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAAACCCAAAGAGUGCGAAAAAGGUGAAAA_AAAGUAA_AAAG .(((((((((...((((((.(((((((....))))))).)))))).....)))))((..(((.((((((............))))....)).)))...))))))................ (-20.28 = -20.72 + 0.44)



| Location | 19,724,746 – 19,724,846 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
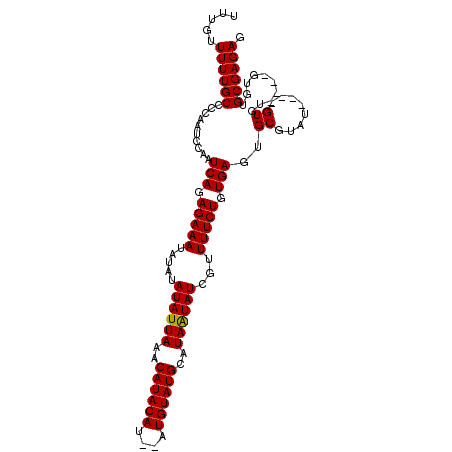
>3L_DroMel_CAF1 19724746 100 + 23771897 UUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAU--AUGUAUGCAUAAUAUCUUUUUCUGUGAGUGCGUAU----GUGU----GUGUGCGAGAG .....((((((.....((......))................((((((((--((((((.((((...........)))).))))))))----))))----))..)))))). ( -22.20) >DroSec_CAF1 34064 104 + 1 UUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAUAUAUGUAUGCAUAAUAUCGUUUUCUGUGAGUGCGUAUAU--GUGU----GUGUGCGAGAG .....((((((.....((......))................((((((((((((((((.((((...........)))).))))))))))--))))----))..)))))). ( -24.60) >DroSim_CAF1 36215 108 + 1 UUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAUAUAUGUAUGCAUAAUAUCGUUUUCUGUGAGUGCGUAUAU--GUGUGUGUGUGUGCGAGAG .....((((((.....((......))................((((((((((((((((((((.(((........)))..))))))))))--))))))))))..)))))). ( -28.70) >DroEre_CAF1 36922 98 + 1 UUUGUUUUUGCCCCAAACCAAUCAGAGAAAAAUAUAUAUUAAACAUACAU--AUGUAUGCAUAGUAUCGUUUUCUGUGAGUGC------GUGUGA----GUGUGCGAGAG .....((((((((((.((((.(((.((((((....((((((..(((((..--..)))))..))))))..)))))).))).)).------)).)).----).).)))))). ( -25.00) >consensus UUUGUUUUUGCCCCAAUCCAAUCAGAGAAAUAUAUAUAUUAAACAUACAU__AUGUAUGCAUAAUAUCGUUUUCUGUGAGUGCGUAU____GUGU____GUGUGCGAGAG .....((((((..........(((.(((((.....((((((..((((((....))))))..))))))...))))).)))..((........))..........)))))). (-17.79 = -17.60 + -0.19)


| Location | 19,724,746 – 19,724,846 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19724746 100 - 23771897 CUCUCGCACAC----ACAC----AUACGCACUCACAGAAAAAGAUAUUAUGCAUACAU--AUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAA ...........----....----....((.((((((((.(((.((((((.((((((..--..)))))).)))))).....))).)))).......))))))......... ( -18.41) >DroSec_CAF1 34064 104 - 1 CUCUCGCACAC----ACAC--AUAUACGCACUCACAGAAAACGAUAUUAUGCAUACAUAUAUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAA .((((((....----....--......))..(((.(((((...((((((.(((((((....))))))).)))))).....))))).))).......)))).......... ( -18.04) >DroSim_CAF1 36215 108 - 1 CUCUCGCACACACACACAC--AUAUACGCACUCACAGAAAACGAUAUUAUGCAUACAUAUAUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAA ...................--......((.((((..((((...((((((.(((((((....))))))).)))))).....))))(((....))).))))))......... ( -18.00) >DroEre_CAF1 36922 98 - 1 CUCUCGCACAC----UCACAC------GCACUCACAGAAAACGAUACUAUGCAUACAU--AUGUAUGUUUAAUAUAUAUUUUUCUCUGAUUGGUUUGGGGCAAAAACAAA .....((...(----(((.((------.((.(((.((((((..(((.((.((((((..--..)))))).)).)))....)))))).))).)))).))))))......... ( -20.70) >consensus CUCUCGCACAC____ACAC____AUACGCACUCACAGAAAACGAUAUUAUGCAUACAU__AUGUAUGUUUAAUAUAUAUAUUUCUCUGAUUGGAUUGGGGCAAAAACAAA .((((((....................))..(((.(((((...((((((.(((((((....))))))).)))))).....))))).))).......)))).......... (-16.75 = -17.00 + 0.25)



| Location | 19,724,846 – 19,724,962 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.13 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
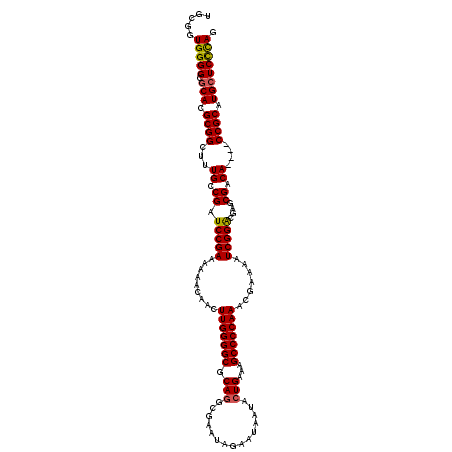
>3L_DroMel_CAF1 19724846 116 + 23771897 UGCGAUGGGGCGCACGCGGCUUUGCCGAUCCGAAAAAACAACUUGGGGCGCAGGAGAAUAGAAUAAUACUGAAAGCCCCAAACGAAAAUCGGGCGAGCGACA----CCGCAUGCUCCCAG .....(((((.(((.((((..((((((.(((((.........(((((((.(((...............)))...))))))).......))))))).))))..----)))).)))))))). ( -47.35) >DroSec_CAF1 34168 120 + 1 UGCGGUGGGGCGCACGCGGCUUUGCCGAUCCGAAAAAACAACUUGGGGCGCAGGCGAAUAGAAUAAUACUGAAAGCCCCAAACGAAAAUCGGACGAGCGACAUGCUCCGCAUGCUCCCAG .....(((((.(((.((((..((((((.(((((.........(((((((.(((...............)))...))))))).......))))))).))))......)))).)))))))). ( -44.45) >DroSim_CAF1 36323 120 + 1 UGCGGUGGGGCGCACGCGGCUUUGCCGAUCCGAAAAAACAACUUGGGGCGCAGGCGAAUAGAAUAAUACUGAAAGCCCCAAACGAAAAUCGGACGAGCGACAUGCUCCGCAUGCUCCCAG .....(((((.(((.((((..((((((.(((((.........(((((((.(((...............)))...))))))).......))))))).))))......)))).)))))))). ( -44.45) >DroEre_CAF1 37020 116 + 1 UGCGCUGGGGCGCACGCGGCUUUGGCGAUCCGAAAAAACAACUUGGGGCGCAAGCGAAUAGAAUAAUACUGAAAGCCCCAAACGAAAAUCGGGCGAGCGACA----CCGCAUGCUCUCAG ....((((((.(((.((((..(((.((.(((((.........(((((((...((..............))....))))))).......)))))))..)))..----)))).))))))))) ( -38.33) >DroYak_CAF1 35050 116 + 1 UGCGGUGGGGCGCACGCGGCUUUGGCGAUCCGAAAAAACAACUUGGGGCGCAGGCGAAUAGAAUAAUACUGAAAGCCCCAAACGAAAAUCGGACGAGCGACA----CCGCAUGAUCCUAG (((((((...(((.(((.......))).(((((.........(((((((.(((...............)))...))))))).......)))))...))).))----)))))......... ( -42.15) >consensus UGCGGUGGGGCGCACGCGGCUUUGCCGAUCCGAAAAAACAACUUGGGGCGCAGGCGAAUAGAAUAAUACUGAAAGCCCCAAACGAAAAUCGGACGAGCGACA____CCGCAUGCUCCCAG .....(((((.(((.((((...((.((.(((((.........(((((((.(((...............)))...))))))).......)))))....)).))....)))).)))))))). (-38.29 = -38.13 + -0.16)

| Location | 19,724,846 – 19,724,962 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -39.26 |
| Energy contribution | -40.50 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19724846 116 - 23771897 CUGGGAGCAUGCGG----UGUCGCUCGCCCGAUUUUCGUUUGGGGCUUUCAGUAUUAUUCUAUUCUCCUGCGCCCCAAGUUGUUUUUUCGGAUCGGCAAAGCCGCGUGCGCCCCAUCGCA .((((.((((((((----(...((.((.((((...(..((((((((...(((...............))).))))))))..).....))))..))))...)))))))))..))))..... ( -45.96) >DroSec_CAF1 34168 120 - 1 CUGGGAGCAUGCGGAGCAUGUCGCUCGUCCGAUUUUCGUUUGGGGCUUUCAGUAUUAUUCUAUUCGCCUGCGCCCCAAGUUGUUUUUUCGGAUCGGCAAAGCCGCGUGCGCCCCACCGCA .((((.((((((((....(((((...((((((...(..((((((((...(((...............))).))))))))..).....)))))))))))...))))))))..))))..... ( -48.16) >DroSim_CAF1 36323 120 - 1 CUGGGAGCAUGCGGAGCAUGUCGCUCGUCCGAUUUUCGUUUGGGGCUUUCAGUAUUAUUCUAUUCGCCUGCGCCCCAAGUUGUUUUUUCGGAUCGGCAAAGCCGCGUGCGCCCCACCGCA .((((.((((((((....(((((...((((((...(..((((((((...(((...............))).))))))))..).....)))))))))))...))))))))..))))..... ( -48.16) >DroEre_CAF1 37020 116 - 1 CUGAGAGCAUGCGG----UGUCGCUCGCCCGAUUUUCGUUUGGGGCUUUCAGUAUUAUUCUAUUCGCUUGCGCCCCAAGUUGUUUUUUCGGAUCGCCAAAGCCGCGUGCGCCCCAGCGCA (((.(.((((((((----(...((....((((...(..((((((((....(((............)))...))))))))..).....))))...))....)))))))))..).))).... ( -37.50) >DroYak_CAF1 35050 116 - 1 CUAGGAUCAUGCGG----UGUCGCUCGUCCGAUUUUCGUUUGGGGCUUUCAGUAUUAUUCUAUUCGCCUGCGCCCCAAGUUGUUUUUUCGGAUCGCCAAAGCCGCGUGCGCCCCACCGCA ...((..(((((((----(...((..((((((...(..((((((((...(((...............))).))))))))..).....)))))).))....))))))))..))........ ( -38.36) >consensus CUGGGAGCAUGCGG____UGUCGCUCGUCCGAUUUUCGUUUGGGGCUUUCAGUAUUAUUCUAUUCGCCUGCGCCCCAAGUUGUUUUUUCGGAUCGGCAAAGCCGCGUGCGCCCCACCGCA ..(((.((((((((....(((((...((((((...(..((((((((...(((...............))).))))))))..).....)))))))))))...)))))))).)))....... (-39.26 = -40.50 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:53 2006