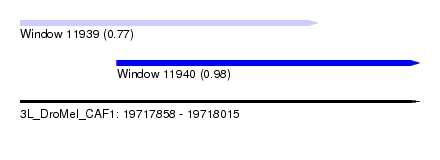
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 19,717,858 – 19,718,015 |
| Length | 157 |
| Max. P | 0.982331 |

| Location | 19,717,858 – 19,717,975 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19717858 117 + 23771897 CUGAC--CUUGAUUUUGCUUUUUAUCGCUCCUGGAUUGGUUCUUUCGUCUGCCUUCAAUUCAACUUCUCGCCGAAACUUUUGGCCGCGGA-AAAGUUGAAUAAACUUGGCCGAAAGGCCA .....--....((((.(((((((..(((...((((((((...............)))))))).......(((((.....))))).)))))-))))).)))).....(((((....))))) ( -31.86) >DroSec_CAF1 26930 108 + 1 CUGAC--CUUGAUUUUGCUUUUUAUCGCUUCUGGAUUGGUUUUU----------UCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGGCCGAAAGGCCA .....--....(((..(((((((.((((...((((((((.....----------)))))))).......(((((.....))))).)))))))))))..))).....(((((....))))) ( -34.90) >DroSim_CAF1 29121 110 + 1 CUGACCCCUUGACCUUUCUUUUUAUCGCUUCUGGAUUGGUUCUU----------UCAGUUCAAAUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGACCGAAAGGCCA .........((.((((((......((((...((((((((.....----------)))))))).......(((((.....))))).))))...(((((.....)))))....)))))).)) ( -26.50) >DroEre_CAF1 29647 113 + 1 CUGAC--CUUGAUUCUGCCCUUUUUCGCUCCUGGGUUGGCUU---CAUCUCUCUUCGGUUCAACUUCUCGCCGA-ACUUUUGGCCGCGGA-AAAGUUUAAUAAACUUGGCCGAAAGGCCA .....--............(((((((((.((.((((((((..---.......(....)...........))).)-))))..))..)))))-))))...........(((((....))))) ( -31.15) >DroYak_CAF1 27675 114 + 1 CAGAC--CUUGAACUUGCUCUUUAACGCUUCUGGGUUGGCUU---CAUCUGUCUUCAGUUAAACUUAUCGCCGAAACUUUUGGGCGCGGA-AAAGUUAAGUAAACUUGGCCGAAAUGCCA ..(((--((.(((...((........))))).)))))(((..---...(((....)))..........((((.((....)).))))(((.-.(((((.....)))))..)))....))). ( -27.00) >consensus CUGAC__CUUGAUUUUGCUUUUUAUCGCUUCUGGAUUGGUUUUU_C_UCU__CUUCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGGA_AAAGUUUAAUAAACUUGGCCGAAAGGCCA ........................((((...((((((((...............)))))))).......(((((.....))))).))))...(((((.....)))))((((....)))). (-20.28 = -20.56 + 0.28)

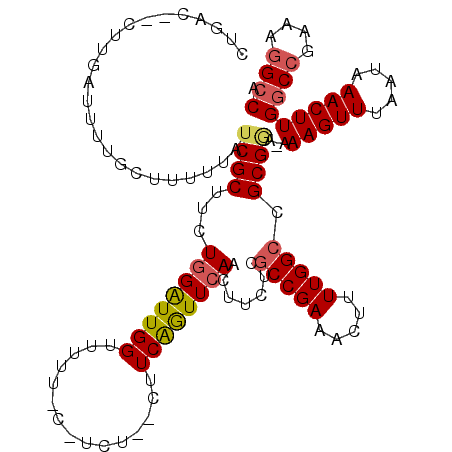
| Location | 19,717,896 – 19,718,015 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.92 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 19717896 119 + 23771897 UCUUUCGUCUGCCUUCAAUUCAACUUCUCGCCGAAACUUUUGGCCGCGGA-AAAGUUGAAUAAACUUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGAUGGGGUAAUUAUUACUUCUG ....(((((.((((((.(((((((((.(((((((.....)))))....))-.))))))))).....(((((....)))))))))...))......))))).((((((.....)))))).. ( -39.30) >DroSec_CAF1 26968 110 + 1 UUUU----------UCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGUUGGGGUAUUUAUUACUUCUG ....----------..((((((.(((((.(((((.....)))))........(((((.....)))))((((....)))))))))))))))...........((((((.....)))))).. ( -32.50) >DroSim_CAF1 29161 110 + 1 UCUU----------UCAGUUCAAAUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGACCGAAAGGCCAGAAGUGAGCUUAAUUGGCGAUGAGGUAUUUAUUACUUCUG ....----------.............(((((((.((((((((((.((....(((((.....)))))...))...))))))))))........))))))).((((((.....)))))).. ( -33.60) >DroEre_CAF1 29685 104 + 1 UU---CAUCUCUCUUCGGUUCAACUUCUCGCCGA-ACUUUUGGCCGCGGA-AAAGUUUAAUAAACUUGGCCGAAAGGCCAGAAUUGAGCUUUAUUGUCGAUGGAGUAUU----------- ..---...((((..(((............(((((-....))))).((((.-((((((((((....((((((....)))))).)))))))))).))))))).))))....----------- ( -32.80) >DroYak_CAF1 27713 116 + 1 UU---CAUCUGUCUUCAGUUAAACUUAUCGCCGAAACUUUUGGGCGCGGA-AAAGUUAAGUAAACUUGGCCGAAAUGCCAGGAGUGUGCUUUAUUGUCGAUUGGGUAUUUAUGACUUCUU .(---(((..((.((((((..(...((..((....((((((((...(((.-.(((((.....)))))..))).....))))))))..))..))...)..)))))).))..))))...... ( -26.90) >consensus UUUU_C_UCU__CUUCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGGA_AAAGUUUAAUAAACUUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGAUGGGGUAUUUAUUACUUCUG ......................((((((((.(((.((((((((((.(((...(((((.....)))))..)))...))))))))))........))).))).))))).............. (-24.34 = -24.94 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:43 2006